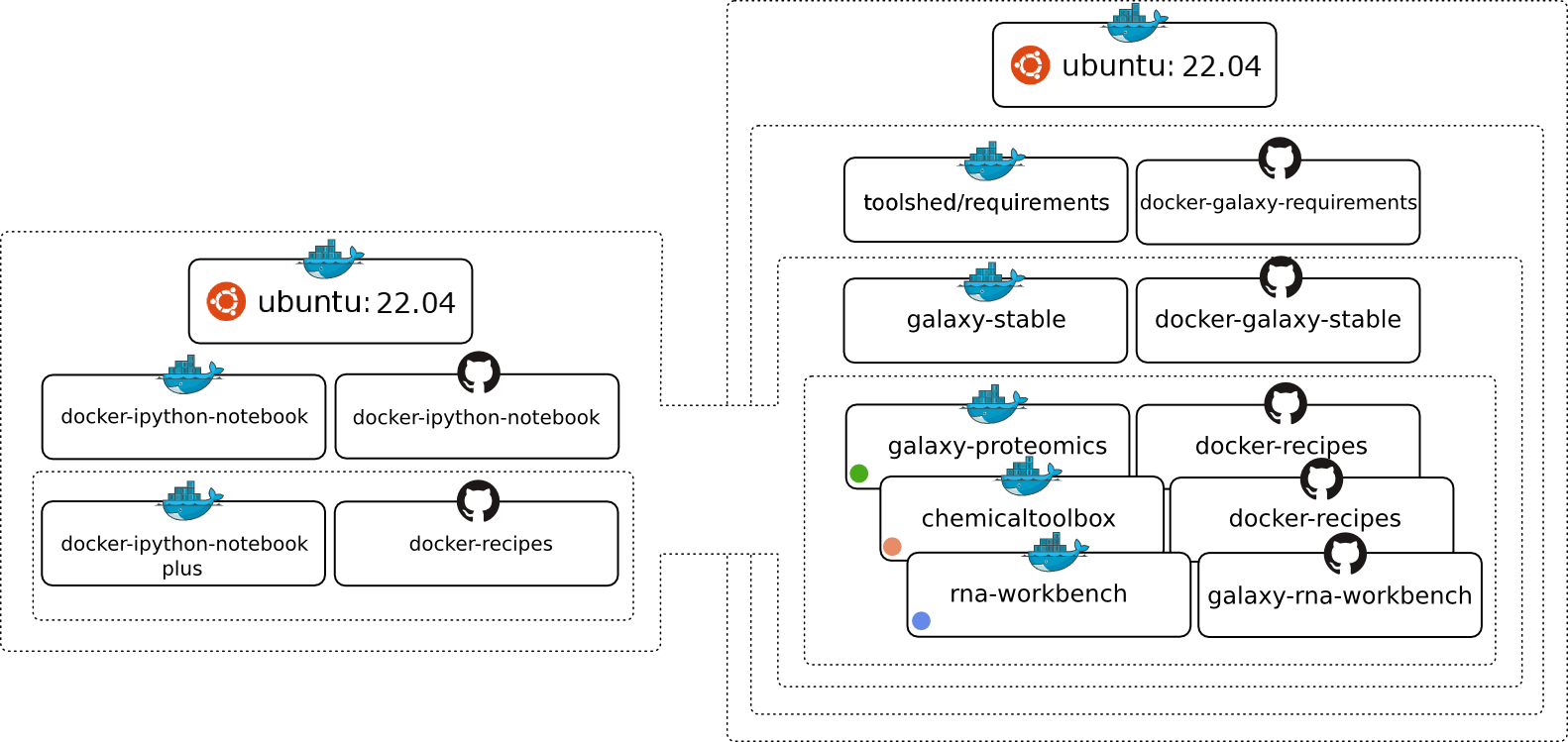
Galaxy Docker
Galaxy Docker provides a ready-to-use Docker-based distribution of the Galaxy platform, enabling users to deploy Galaxy quickly and reproducibly on local machines, servers, or cloud environments. It offers a convenient way to start a fully functional Galaxy instance without manual installation of dependencies, making it well suited for testing, training, and portable analysis setups.
Key benefits
- Fast and reproducible deployment of a Galaxy instance using Docker
- No complex local installation—dependencies and configuration are packaged into the container
- Portable setup for local use, servers, and cloud/HPC-adjacent environments
- Useful for training courses, workshops, demos, and prototyping
- Open-source and maintained in a public repository
Applications
- Rapid setup of Galaxy for teaching, workshops, and hands-on tutorials
- Testing Galaxy configurations, tools, or workflows in an isolated environment
- Providing a portable Galaxy environment for teams or collaborations
- Quick prototyping of analysis environments prior to production deployment
Intended use
Galaxy Docker is intended for bioinformatics users, trainers, and service operators who want to deploy Galaxy quickly for temporary or reproducible environments. It is particularly suited for users who prefer a containerized approach for setup and maintenance, or who need a reliable Galaxy instance for training and testing scenarios.
Help us improve our de.NBI services – take 2 minutes to fill in our survey. Thank you!
Galaxy Docker on GitHub