J. Kalinowski, A. Sczyrba, Bielefeld University
Service center: Bielefeld-Giessen Resource Center for Microbial Bioinformatics – BiGi
COVID-19, caused by the novel coronavirus SARS-CoV-2 is a worldwide threat to healthcare and to economy. In improved spatio-temporal resolution of the pandemy by using the mutation patterns in the virus‘ RNA genome is crucial to follow infection chains, detect hotspots and to decide on the efficiency of countermeasures.
In close cooperation with a local clinic in Bielefeld, Germany, we have sequenced several SARS-CoV-2 genomes and detected novel mutational signatures that help in dissecting the outbreak. Currently, we aim at integrating all clinics and testing labs in the region, to sequence several hundred cases or a fine grained map of the region including information on locations of patients and mutational signatures of their SARS-CoV-2 viruses. This map will be gradually expanded over time in order to assess pre-lockdown sezenarios and those appearing at later stages of the outbreak, delivering critical data for a long-term surveillance. We also aim to integrate our local epidemiology data with national and international (https://leoss.net/) efforts in the same direction.
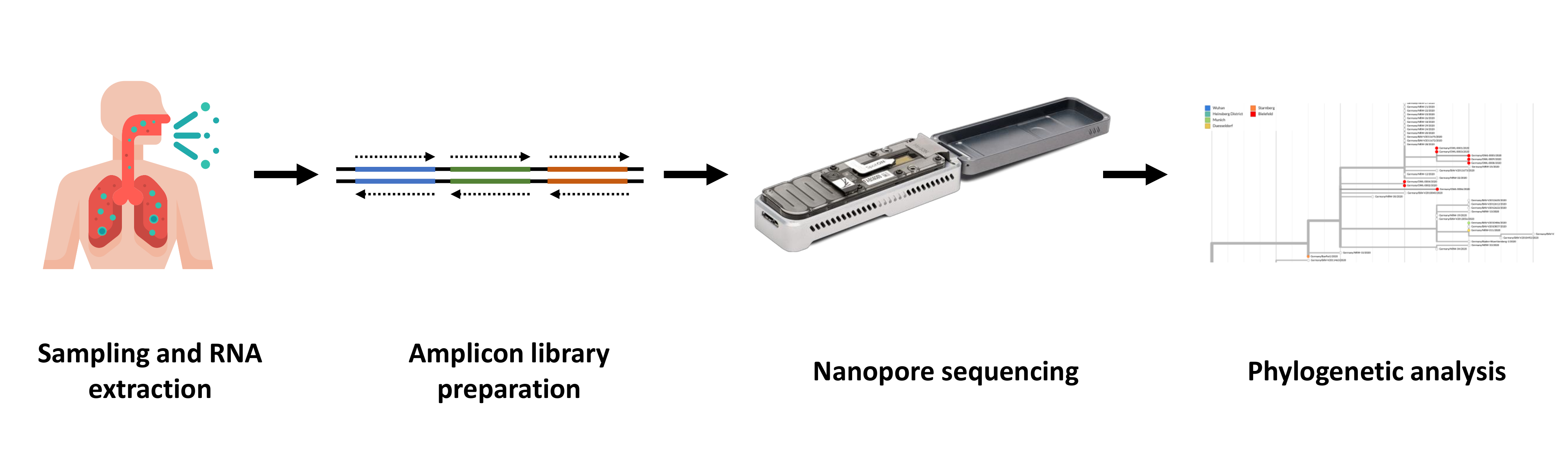
Sequencing has been performed by adapting the ARTIC protocol for nanopore sequencing (https://artic.network/ncov-2019) that provided primer sequences as well as lab methods and bioinformatics tools for generating full-length virus sequences from nucleic acids that have been isolated for the RT-PCR diagnostic procedure. Nanopore sequencing of amplicons that were generated in multiplex PCRs has proven to be precise enough to unequivocally call single nucleotide polymorphisms (SNP) for dissecting virus subtypes. SNP patterns have been validated successfully by Sanger sequencing of PCR amplicons or by complete transcriptome sequencing using the Illumina method.
Sequences have also been used to reconstruct global phylogenies on the de.NBI cloud infrastructure, proving that at least one genotype has hitherto only been found in Bielefeld. Data will be uploaded into international nucleic acid sequence repositories following the FAIR principle. Additionally, the BIGI service center will expand its established nanopore software training course by a hands-on lab module with a focus on SARS-CoV-2 sequencing.