R. Eils, HiDiH Berlin
Service center: Heidelberg Center for Human Bioinformatics – HD-HuB
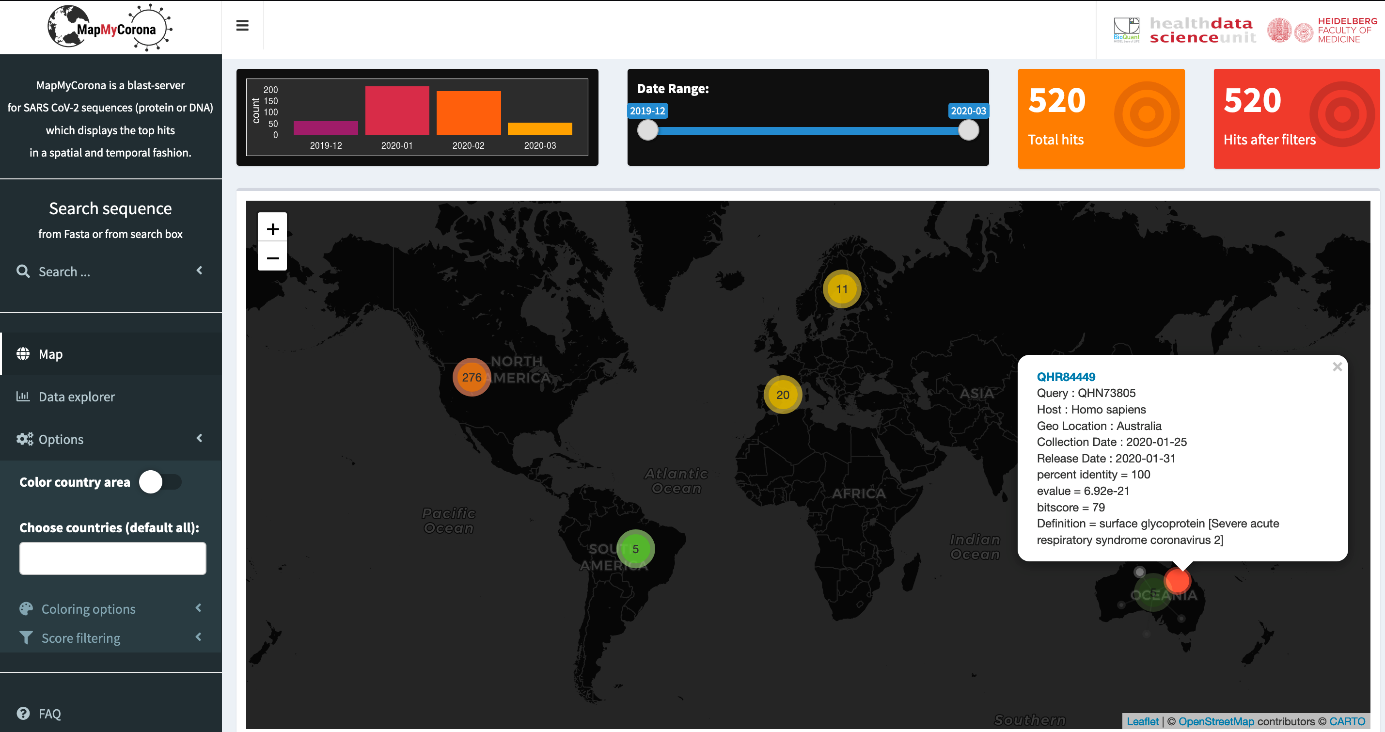
The corona pandemics has shown an impressive speed of spreading throughout the world. Within a few weeks, most countries have reported infections, with high rates of new daily infections. An interesting question is to what extend the viral sequence has evolved during this spreading: are there any signs of adaptation of the viral to new hosts, or could there be multiple strains involved in this global pandemic? In order to tackle these questions, the viral sequences are being sequenced by labs around the world, which report the viral sequences from diverse patient samples. MapMyCorona uses a central database of viral sars-cov-2 sequences to provide an easy way to display sequence similarity or alterations in a query sequence, compared with the already available sequences. We implemented a blast server, which displays the top hits of a query sars-cov-2 sequences to other sequenced corona virus on a world map, thus enabling a direct interpretation of the best matching hits. The tool also provides information on the percent similarity, and information on the target sequence, and can thus be used to follow the temporal and spatial dissemination of a viral sars-cov-2 strain.
For further information, visit https://hdsu-bioquant.shinyapps.io/mapmycorona/