H. Erfle, Ruprecht Karl University of Heidelberg
Service center: Heidelberg Center for Human Bioinformatics – HD-HuB
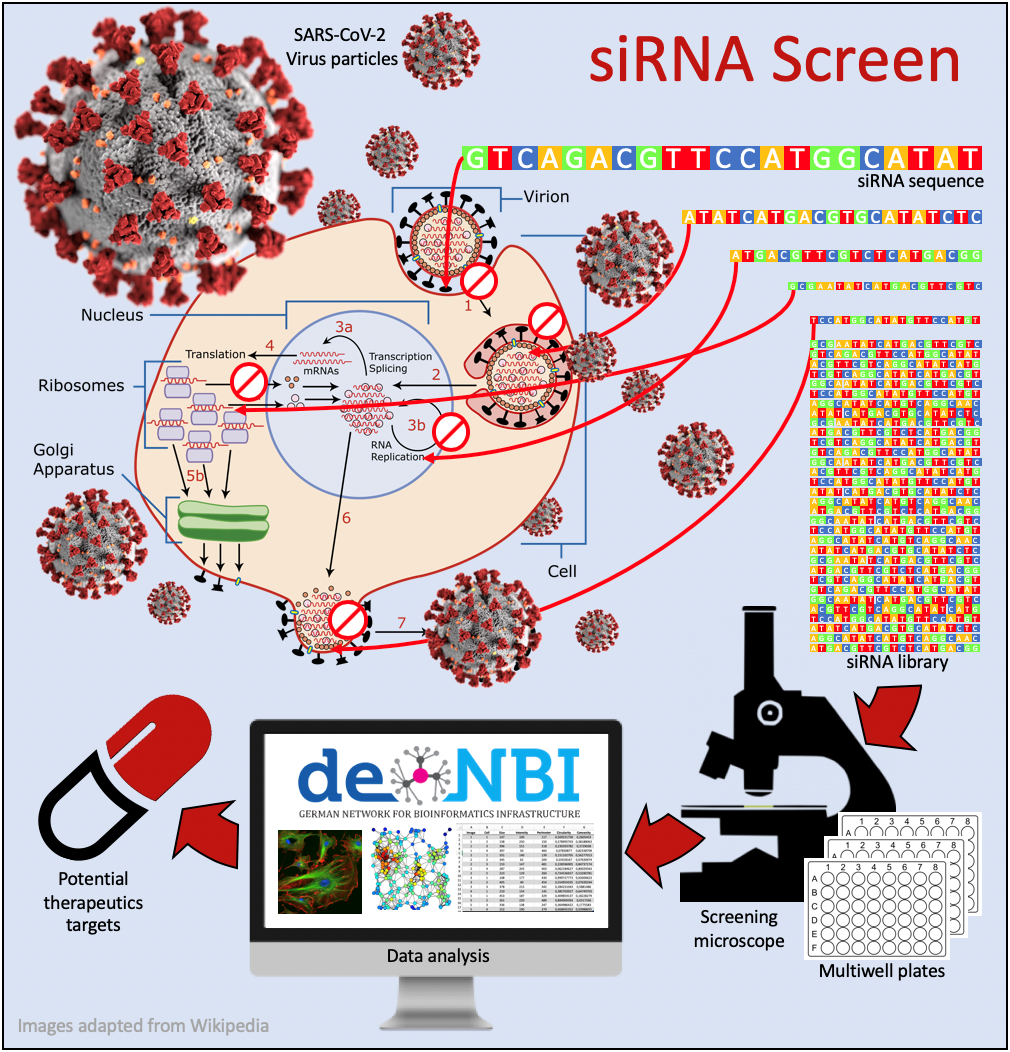
End of 2019 a novel virus was reported later identified by blood samples, respiatory swab and next generation sequencing as a Coronavirus and termed SARS-CoV-2, inducing the infectious disease COVID19. Currently, the exact mechanisms how the novel SARS-CoV-2 virus is able to enter human cells, replicate there and exit again highly amplified are not well understood. Like for any virus, these processes are highly dependent on genes or proteins inherent to the infected host.
Together with scientists from the University Hospitals in Frankfurt and Jena, we are planning to conduct a systematic screen for those host factors which are potentially affectable by drugs and drug combinations. For this, we transfect human cells from an immortalized model cell line with a library of RNA fragments which are capable of suppressing the transcription of a specific gene into a protein, thus knocking the copies of this protein temporarily down inside the cells. Each of the thousands of RNA fragments (short interfering RNA, siRNA) is tested in a separate experiment, for each gene there are three different siRNA sequences tested. In parallel, the cells are infected with SARS-CoV-2 and their behavior is observed with a high-throughput microscope. All these processes are, once set up, fully automated. The microscopic images are analyzed for specific alterations of the cells with image processing pipelines which we also offer as a service in de.NBI. It is quantified how these cellular alterations occur upon viral infection (systematic phenotyping) or if the cells stay healthy. If they stay healthy, it is a strong indicator that the currently knocked down protein was essential for the virus either to enter, to replicate in or to exit the cells. The results of these screening experiments are further analyzed in order to identify protein networks and pathways crucial for host-virus interactions of SARS- CoV-2.
In our screen, we concentrate on siRNA against genes potentially affectable by drugs, so that we hope to identify drugs or drug combinations that are effectively preventing or stopping the infection of healthy cells with SARS-CoV-2.
For further information, visit:
http://www.bioquant.uni-heidelberg.de/technology-platforms/viroquant-cellnetworks-rnai-screening- facility/screening-core-facility.html
For further information about systematic phenotyping tools, visit https://www.hd-hub.de/services/services-systematic-phenotyping/