R. Backofen, Albert Ludwigs University Freiburg
Service center: RNA Bioinformatics Center – RBC
Public archives and databases, like ENA, SRA, are key to ensuring read sequences and datasets are ideally stored long-term and adhering to the FAIR (Findable, Accessible, Interoperable, Reusable) principles. For more than 15,000 users of the European Galaxy server (https://usegalaxy.eu) we have integrated special connectors to those public archives to easily provide scientists with those data in Galaxy and our clouds. Besides accessing data, keeping track of the latest data is time-consuming and not straightforward due to the rapid data growth. Our Galaxy Gateway is designed for downloading and analysing all available (several thousand) sequences of SARS-CoV-2 that are currently published in those data archives. To overcome the issues of tracking the latest datasets as well as to provide a quick turn-around in analysing the latest sequences, we have created a collection of identifiers of relevant sequence-datasets that we update daily in an automatic way. We also mirror all publicly available COVID-19 related datasets in Galaxy to ease the access and reduce analysis time. To facilitate the COVID-19 research, we also continuously integrate the recent SARS-CoV-2 reference genome and create optimised indices to access it from all related tools. After provision of these key features, the European Galaxy server has recently experienced an increased usage especially in COVID-19 related research. In March 2020, the European Galaxy server processed 400,000 jobs, and in April already 500,000 jobs. In terms of data, 140 TB were uploaded in April or created by our users. The hardware underlying the Galaxy service is provided by the “Federal Ministry of Education and Research” of Germany, which is supporting the de.NBI-cloud; the University of Freiburg, which offered 2000 additional cores to the Galaxy infrastructure to fight the pandemic; and a global distributed compute network with contributions from Finland, Belgium, UK, Italy, Spain, Norway, Portugal, and Australia. Specialised tools, particularly in the field of long-read sequencing and drug design, have advanced requirements, e.g. GPUs. With the help of the University of Freiburg and colleagues from the UK, we were able to offer GPUs to all researchers within only a week to accelerate their research during the COVID-19 pandemic.
For further information visit the Galaxy project website: https://covid19.galaxyproject.org
This work was published: https://doi.org/10.1101/2020.02.21.959973
To access the de.NBI cloud visit de.NBI Cloud under https://www.denbi.de/cloud, to access the European Galaxy server please visit https://usegalaxy.eu
|
|
|
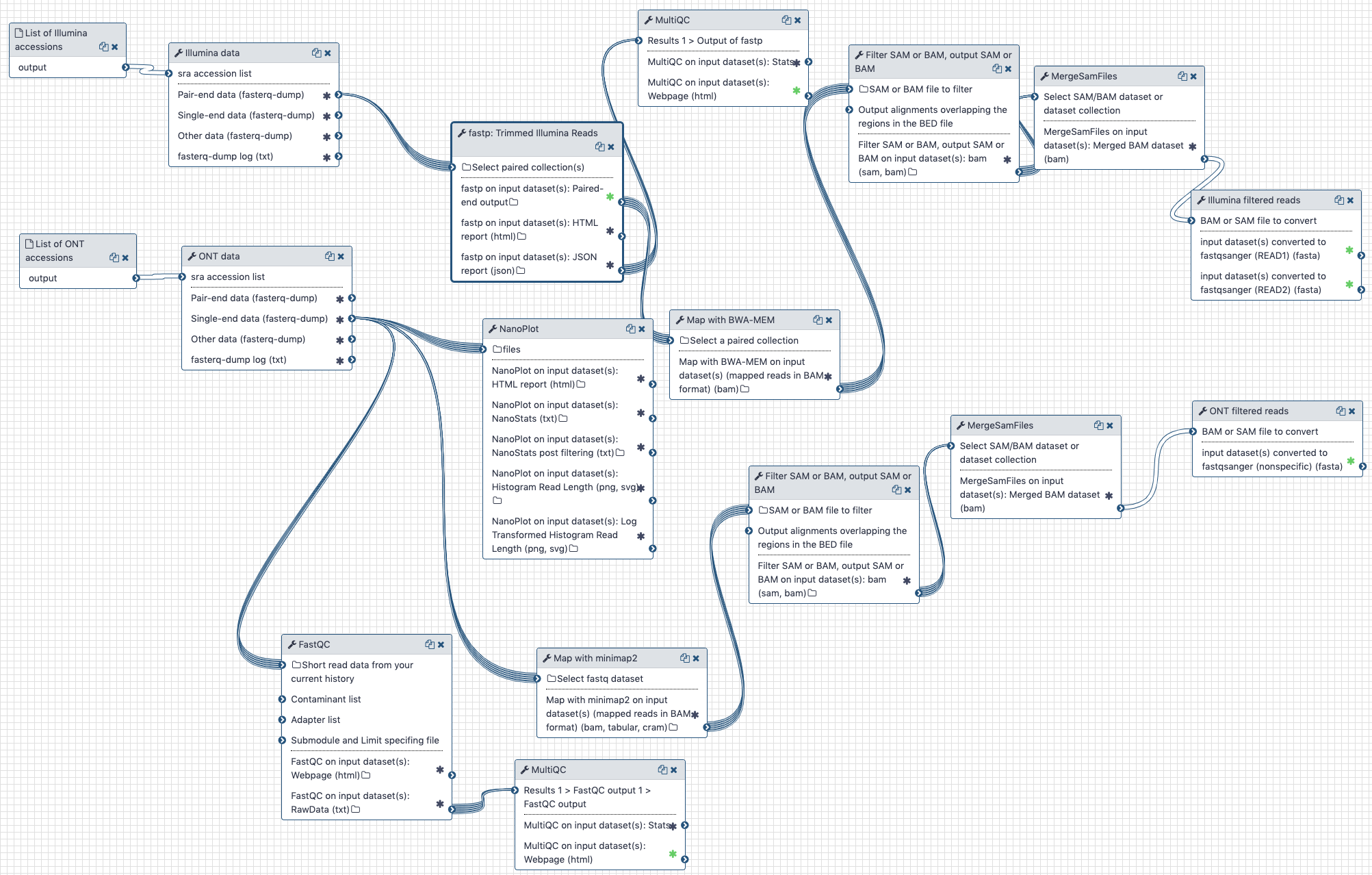
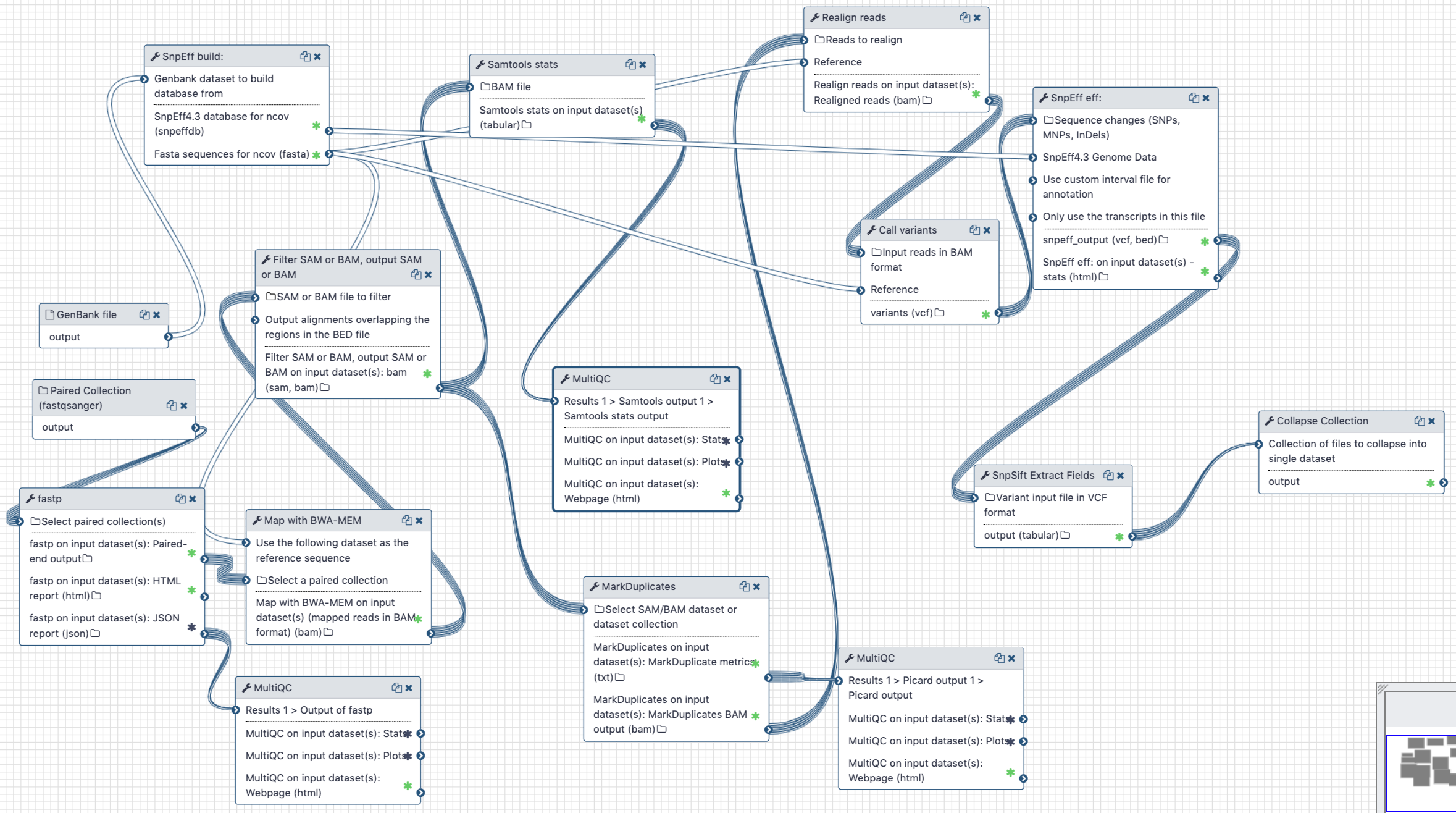
Figure: Example workflows for pre-processing of SARS-CoV-2 short-read and long-read sequences (left) and analysis of paired-end Illumina reads (right).