A. Akalin and N. Rajewsky, Max Delbruck Center for Molecular Medicine, Berlin
Service center: RNA Bioinformatics Center – RBC
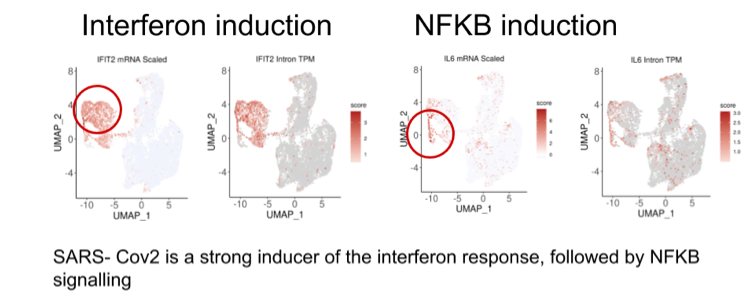
The coronavirus disease 2019 (COVID-19) pandemic, caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is an ongoing global health threat with more than two million infected people since its emergence in late 2019. Detailed knowledge of the molecular biology of the infection is indispensable for the understanding of the viral replication, host responses, and disease progression. In order to understand these aspects, we teamed up with the labs of Prof. Markus Landthaler from MDC and Prof. Christian Drosten from Charite. The experimental team provided the gene expression profiles of SARS-CoV and SARS-CoV-2 infections in three human cell lines (H1299, Caco-2 and Calu-3 cells), using bulk and single-cell transcriptomics. We have analyzed those raw datasets using our robust and reproducible PiGx pipeline (https://bioinformatics.mdc-berlin.de/pigx/). In addition, specially developed downstream analysis workflow is used on single-cell RNA-seq data to identify transcriptome dynamics during infection and to identify potential drug target targets. These and additional analyses in the single-cell space revealed that canonical interferon-stimulated genes such as IFIT2 or OAS2 were broadly induced. In addition, the temporal resolution of transcriptional responses suggested interferon regulatory factors (IRFs) activities precede that of nuclear factor-κB (NF-κB). Lastly, we identified heat shock protein 90 (HSP90) as a protein relevant for the infection and a potential drug target. Inhibiting this protein decreased the viral replication.