| Issue 02/2022 |
 Establishment of a Community of Practice for DM/DS Trainers Establishment of a Community of Practice for DM/DS Trainers
|
|
Research Data management refers to the handling of research data before, during, and after a research activity. To ensure that all relevant aspects of data management are considered from the very beginning on, more and more research organisations and institutes require their researchers to develop data management plans (DMPs).
The scientific community’s rising demand to provide good research data management induced the idea to establish a Community of Practice for Data Management/Data Stewardship (DM/DS) Trainers. This Community of Practice (CoP) is intended for all life scientists who are interested and involved in DM/DS training and who would like to participate in developing, improving and expanding training materials encouraging the exchange of expertise. The goal is to specify common standards in data management training and to set a universal base level of knowledge with regards to best practices through training in data management.
In the framework of the European project ELIXIR-CONVERGE, a first Training Community kick-off for DM/DS took place in March 2022, with the aim to discuss together with the participants questions related to the organization of this community of practice, in order to then jointly shape the structure of this community. Two external speakers gave great lectures on examples of national DM/DS communities involved in training:
- NFDI project in Germany held by Daniel Tschink (NFDI4Biodiversity, GFBio)
- Data Science Training in Health and Biosciences (DaSH) project presented by Robert Andrews (Cardiff University, UK)
In a subsequent productive discussion round with about 90 participants from all over the world, the goals of the CoP were discussed, how these can be achieved, and which communication channels should be used for this community network. As next community events several hackathons will take place dealing with the topics: DMP, DM/DS Basics, FAIR and Reproducibility. This hackathon series aims to generate a comprehensive set of training materials that are as all-encompassing as possible and taht can be used as a basis for various formats of DM training courses. In this context, ELIXIR Germany has already organised a hackathon series in which a generalised slide deck on the topic of DMP was created. Further hackathons will follow for finalisation.
If you would like to be part of this Community of Practice for Data Management/Data Stewardship Trainers and are interested in using this chance for networking just contact us: This email address is being protected from spambots. You need JavaScript enabled to view it.
|
 “Boost your Data Management Planning” workshops at ECCB & GCB 2022 “Boost your Data Management Planning” workshops at ECCB & GCB 2022
|
|
Have you ever wondered, what research data management really is? Why is it so important? Then de.NBI / ELIXIR-DE has created the perfect training course for you that will take place in September at GCB (5.9.) and ECCB (18.9) 2022!
More and more funders require the establishment of research data management plans for their grants. Over the course of just three hours, you will get the chance to learn how to transfer research project proposals into proper Data Management Plans (DMPs). The data management experts Helena and Daniel (CONVERGE WP2) will guide you through the basics of research data management, the dos and don’ts and how to improve the management of the data produced in research projects.
After a short introduction to the research data life cycle and the FAIR data principles, we will explore in multiple hands-on sessions what a data management plan (DMP) is. We will sink our teeth into components, language, software and examples of DMPs. In light of the FAIR principles, we will evaluate possible problems and solutions and self-assess a drafted DMP for our own projects. See you in Halle (Saale) or Sitges!
|
 Meet ELIXIR Germany at 2022 All Hands Meeting in Amsterdam Meet ELIXIR Germany at 2022 All Hands Meeting in Amsterdam
|
|
This year’s ELIXIR All Hands meeting, the eighth time in a row, will take place from 7-10 June 2022 in Amsterdam and bring together members of the ELIXIR community from across the ELIXIR Nodes. ELIXIR Germany contributes to the organization and presentation of numerous workshops and talks:
Plenary talk:
- Delivering ELIXIR’s large-scale projects (Beatriz Serrano-Solano) – 09.06.2022; 16:00 – 17:20
Mini-Symposia:
- Research Data Management (Ulrike Wittig, Sebastian Beier, Daniel Wibberg, Irena Maus) – 08.06.2022; 09:00 – 10:30
- ELIXIR Communities - Connectedness (Julian Uszkoreit) – 08.06.2022; 11:00 – 12:30
- COVID-19 (Beatriz Serrano-Solano, Björn Grüning) – 08.06.2022; 14:00 – 15:30
Workshops:
- AI-powered data analysis within the European Galaxy (Beatriz Serrano-Solano) – 07.06.2022; 11:00 – 12:30
- Run your own workflow on GA4GH-powered ELIXIR cloud infrastructures (Björn Grüning, S. Twardziok) - 07.06.2022; 14:00 - 15:30
- Development and strengthening of links between the Plant Sciences Community and the ELIXIR Data, Interoperability and Training Platforms Workshop (Sebastian Beier), - 07.06.2022; 11:00 - 12:30
- Establishing a roadmap for the Communities towards a sustainable co-production model with the Training Platform (Bérénice Batut) - 07.06.2022; 14:00 - 15:30
- Earth Biodiversity and Galaxy Workshops (Bérénice Batut, Beatriz Serrano-Solano, Björn Grüning) - 07.06.2022; 14:00 - 15:30
- Science driven improvements to tools ecosystems Workshop (Björn Grüning) - 09.06.2022; 11:00 - 12:30
- ELIXIR Tools Platform Ecosystem: Benchmarking reference workflows in OpenEBench through practical experiences Workshop (Björn Grüning) - 09.06.2022; 14:00 - 15:30
- FAIR in single cell omics; Workflows and Benchmarking Workshop (Naveed Ishaque, Björn Grüning) - 09.06.2022; 14:00 - 15:30
- Identification, collection and review of datasets for the assessment and development of supervised machine learning methods for biomedical applications (Sucheta Ghosh) - 09.06.2022; 14:00 - 15:30
Poster:
- Orchestrate your Nextflow and Snakemake runs with WESkit. (Kaunitz et al.)
- SimpleVM–a simple to use cloud computing environment for the life sciences (Hoffmann et al.)
- The new de.NBI/ ELIXIR-DE Training Platform in 2022 (Wibberg et al.)
- Establishment of a Community of Practice for DM/DS Trainer (Schnitzer et al.)
|
 News from de.NBI and ELIXIR-DE Office News from de.NBI and ELIXIR-DE Office
|
Team of ELIXIR-DE Officers is growing
ELIXIR Germany has established the ELIXIR Officers Team, which supports node-specific activities at the different service units and assists the ELIXIR-DE Hub in fulfilling its tasks. At the moment, German activities are coordinated by seven officers specialized in various bioinformatics topics and working within their fields of expertise in ELIXIR.
 We now extend a warm welcome to Michael R. Crusoe, who started his position as ELIXIR-DE Officer on April 1, 2022. Michael is a one of the Common Workflow Language co-founders and the CWL project lead. He is also a Software Carpentry instructor and Debian software packager. In addition to his employment at ELIXIR-DE, Michael is a member of the Dutch ELIXIR Node where he actively contributes to the ELIXIR Interoperability & Compute Platforms. He is also a part-time PhD student at VU Amsterdam on the topic of workflow standards in practice. Originally from Phoenix, Arizona, USA; Michael is happy to call Berlin, Germany his home after previously living in Lithuania, Romania, and Belgium. We now extend a warm welcome to Michael R. Crusoe, who started his position as ELIXIR-DE Officer on April 1, 2022. Michael is a one of the Common Workflow Language co-founders and the CWL project lead. He is also a Software Carpentry instructor and Debian software packager. In addition to his employment at ELIXIR-DE, Michael is a member of the Dutch ELIXIR Node where he actively contributes to the ELIXIR Interoperability & Compute Platforms. He is also a part-time PhD student at VU Amsterdam on the topic of workflow standards in practice. Originally from Phoenix, Arizona, USA; Michael is happy to call Berlin, Germany his home after previously living in Lithuania, Romania, and Belgium.
https://www.denbi.de/team-elixir-officers/
|
New ELIXIR Germany Deputy Technical Coordinator
 Sebastian Jaenicke took over his new position as Deputy Technical Coordinator in ELIXIR-DE. He succeeds Marius Dieckmann in this function, who is now employed as an NFDI software developer and cloud architect at Giessen University. Sebastian is member of the Bielefeld-Giessen (BiGi) Bioinformatics Resource Center and brings in a lot of experience field of microbial bioinformatics. His research is focusing on the design and further development of a flexible and extensible client/server-framework for the management and analysis of metagenomic datasets, also known as MGX (https://mgx-metagenomics.github.io/) Sebastian Jaenicke took over his new position as Deputy Technical Coordinator in ELIXIR-DE. He succeeds Marius Dieckmann in this function, who is now employed as an NFDI software developer and cloud architect at Giessen University. Sebastian is member of the Bielefeld-Giessen (BiGi) Bioinformatics Resource Center and brings in a lot of experience field of microbial bioinformatics. His research is focusing on the design and further development of a flexible and extensible client/server-framework for the management and analysis of metagenomic datasets, also known as MGX (https://mgx-metagenomics.github.io/)
We welcome Michael and Sebastian in our team and look forward to a successful cooperation!
https://www.denbi.de/elixir-de
|
ELIXIR-DE Industry Officer
 Nils-Christian Lübke from the de.NBI administration office was appointed by the CCU to represent ELIXIR-Germany’s industry liaison efforts as the new “ELIXIR-Germany Industry Officer”. Since 2022, Nils took over the role of the de.NBI Industrial Forum Coordinator from Manuel Wittchen. The de.NBI Industrial Forum provides networking opportunities at the interface between Academia and Industry. His responsibilities in this position already covered connectivity towards ELIXIR. Nils-Christian Lübke from the de.NBI administration office was appointed by the CCU to represent ELIXIR-Germany’s industry liaison efforts as the new “ELIXIR-Germany Industry Officer”. Since 2022, Nils took over the role of the de.NBI Industrial Forum Coordinator from Manuel Wittchen. The de.NBI Industrial Forum provides networking opportunities at the interface between Academia and Industry. His responsibilities in this position already covered connectivity towards ELIXIR.
Now, in his new role as “ELIXIR-Germany Industry Officer”, Nils will also be representing the industrial activities of ELIXIR-Germany within the ELIXIR network. The ELIXIR-Germany Industry Officer is meant to act as a contact person for Industry partners and ELIXIR-Germany members.
Feel free to reach out to Nils: https://www.denbi.de/administration-office-ao-menu/
|
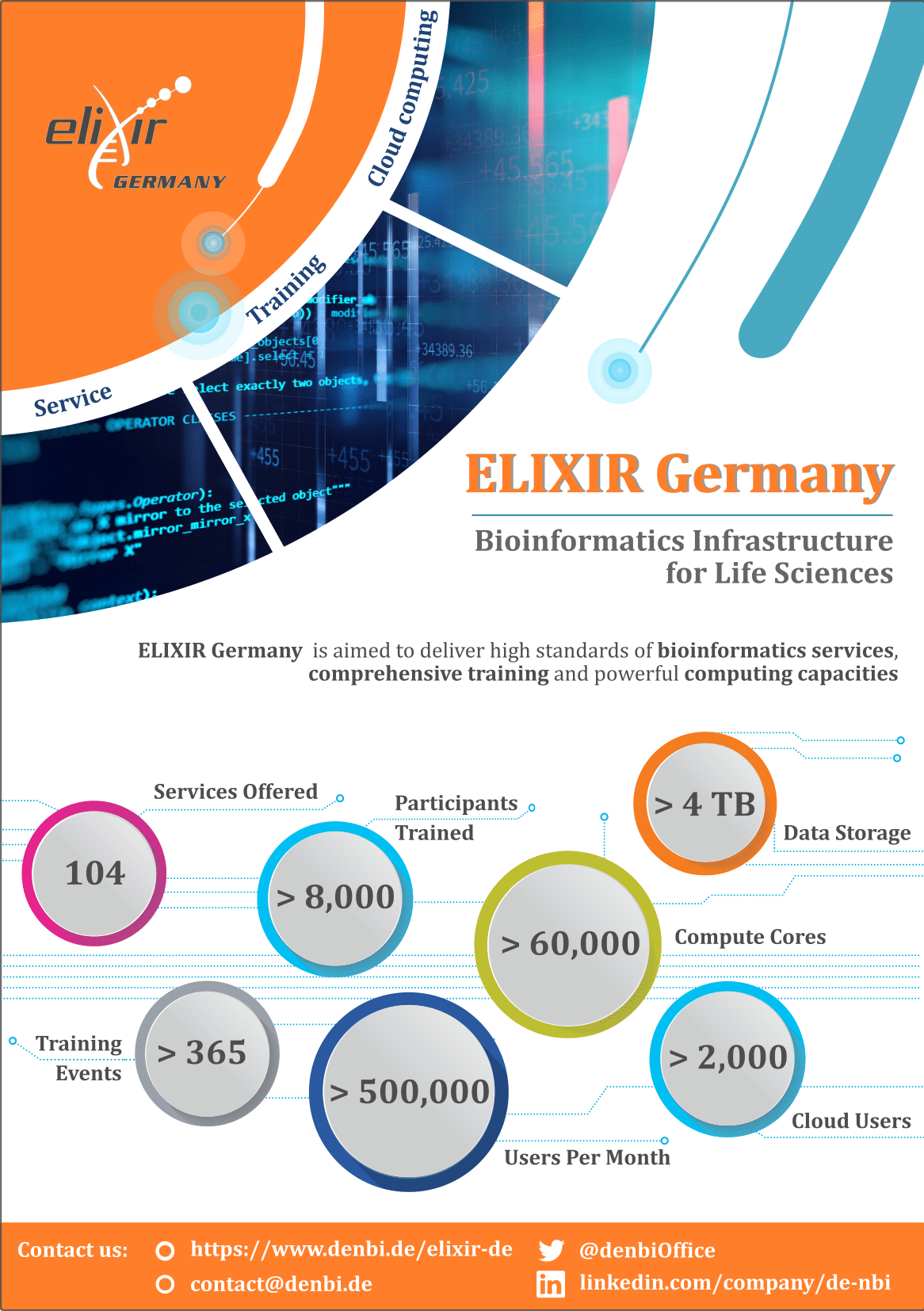
New ELIXIR Germany Impact Flyer
ELIXIR Germany is delighted to present a new impact flyer, which summarizes the most important facts and figures about ELIXIR Germany services at a glance. For more information about ELIXIR Germany please visit the website.
|
 News on the de.NBI cloud News on the de.NBI cloud
|
de.NBI Cloud - Life Science AAI Migration
On 11 April 2022, the de.NBI Cloud Portal and Cloud Compute Centers migrated from ELIXIR AAI as their authentication and authorization provider to the new EOSC Life Science AAI provider. The migration required a short downtime of the Cloud Compute Centers on April 11th and a longer downtime of the de.NBI Cloud portal from April 11th - 12th. Users were automatically migrated to the new provider and for most, the migration caused no issues.
Please note that support for social providers (Google, Orcid ...) does not convey institutional membership information and thus may lead to issues and longer review times of your project applications. Should you note any issues with authentication, please contact either us at This email address is being protected from spambots. You need JavaScript enabled to view it. or preferably the Life Science AAI support at This email address is being protected from spambots. You need JavaScript enabled to view it.
ELIXIR Compute Platform Face 2 Face Meeting in Berlin
On 25-26 April 2022, the ELIXIR Compute Platform Face 2 Face Meeting took place in Berlin with around 24 on-site and up to 10 remote participants. The meeting started with a welcome and overview of ELIXIR Germany’s Cloud Activities by Dr. Nils Hoffmann, ELIXIR Germany’s Cloud Platform Coordinator. After "A brief history of the ELIXIR Compute Platform" by one of the initial founders of the ELIXIR Compute Platform, Ludek Matyska (ELIXIR CZ), Jonathan Tedds (ELIXIR Hub) presented the current platform portfolio. In further sessions, the latest status of ELIXIR Compute Platform tasks was presented by the respective task leads and contributors, including work packages and associated deliverables. Topics such as the ELIXIR to Life Science AAI migration were presented and discussed, as well as involvement in EOSC Life and GA4GH related activities. Day two of the meeting was kicked off by Harald Wagener (ELIXIR DE, BIH@Charité) who presented "HEALTH-X dataLOFT: A Gaia-X Health Data Space for Europe".
The successful two-day meeting continued with updates by the different compute platform tasks and ended on Tuesday afternoon with discussions on future directions and objectives for the ELIXIR Compute Platform.
|
 News on Services News on Services
|
|
Numerous improvements of de.NBI services have been reported in the last quarter:
usegalaxy.eu: Blog by Björn Grüning: Towards a sustainable storage, enabling co-financing of public infrastructure
BacDive:
License was made even more permissive with the Creative Commons Attribution 4.0 International License.
Update on SILVA taxonomy:
new release 138. SILVA’s most recent taxonomic work has focused on the maintenance of the resource, including the rectification of reported errors and the incorporation of changes coming from external taxonomy resources. For more details, please check the SILVA taxonomy documentation.
EggNOG:
EggNOG-mapper web is now using eggNOG-mapper 2.1.7, mainly (hopefully) fixing an issue with GFF positions.
Carlos P Cantalapiedra, Ana Hernández-Plaza, Ivica Letunic, Peer Bork, Jaime Huerta-Cepas, eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale, Molecular Biology and Evolution, Volume 38, Issue 12, December 2021, Pages 5825–5829, https://doi.org/10.1093/molbev/msab293
Copasi:
The COPASI team announces the immediate availability of the stable release COPASI 4.35 (Build 258). More information can be found in the changelog.
Jonas Förster, Frank T. Bergmann, Jürgen Pahle, CoRC: the COPASI R Connector, Bioinformatics, Volume 37, Issue 17, 1 September 2021, Pages 2778–2779, https://doi.org/10.1093/bioinformatics/btab033
|
 Recent Publications by de.NBI / ELIXIR-DE partners Recent Publications by de.NBI / ELIXIR-DE partners
|
- Alfred Pühler - "Das Deutsche Netzwerk für Bioinformatik-Infrastruktur de.NBI" in Innovationsradar Klimawende Kompetenznetzwerk Umweltwirtschaft NRW
- V-F Schumann, ... R Wurmus, ..., B Uyar, V Franke, ..., N Rajewsky, M Landthaler, A Akalin; SARS-CoV-2 infection dynamics revealed by wastewater sequencing analysis and deconvolution medRxiv 2021.; https://doi.org/10.1101/2021.11.30.21266952
- J Breuer, ... A Goesmann et al. What goes around comes around: artificial circular RNAs bypass cellular antiviral responses, Molecular Therapy - Nucleic Acids, 2022, ISSN 2162-2531, https://doi.org/10.1016/j.omtn.2022.04.017.
- Y Ren, ...., A Goesmann, et al.; Prediction of antimicrobial resistance based on whole-genome sequencing and machine learning, Bioinformatics, Volume 38, Issue 2, 15 January 2022, Pages 325–334, https://doi.org/10.1093/bioinformatics/btab681
- R M Waterhouse, A-F Adam-Blondon, ..., C Quast, FO Glöckner, ..., A Nunes-Jorge, NP Willassen, J Lanfear, Recommendations for connecting molecular sequence and biodiversity research infrastructures through ELIXIR. F1000Research 2021, 10(ELIXIR):1238, doi.org/10.12688/f1000research.73825.1
- M Akgün, N Pfeifer, O Kohlbacher, Efficient privacy-preserving whole-genome variant queries, Bioinformatics, Volume 38, Issue 8, 15 April 2022, Pages 2202–2210, https://doi.org/10.1093/bioinformatics/btac070
- S Bray, ..., R Backofen, B Grüning et al. Galaxy workflows for fragment-based virtual screening: a case study on the SARS-CoV-2 main protease. J Cheminform 14, 22 (2022). https://doi.org/10.1186/s13321-022-00588-6
- F Meyer, ..., K Reinert, ..., G Zeller, ..., A Sczyrba & AC McHardy; Critical Assessment of Metagenome Interpretation: the second round of challenges. Nat Methods 19, 429–440 (2022). https://doi.org/10.1038/s41592-022-01431-4
- M Palmblad, S Böcker, S Degroeve, O Kohlbacher et al., Interpretation of the DOME Recommendations for Machine Learning in Proteomics and Metabolomic., Proteome Res. 2022, 21, 4, 1204–1207, https://doi.org/10.1021/acs.jproteome.1c00900
- O Alka, ..., O Kohlbacher & Hannes L. Röst, DIAMetAlyzer allows automated false-discovery rate-controlled analysis for data-independent acquisition in metabolomics. Nat Commun 13, 1347 (2022). https://doi.org/10.1038/s41467-022-29006-z
- P Brack; ...; F Coppens B Grüning, I Eguinoa, P Ewels, C Goble, Ten simple rules for making a software tool workflow-ready PLOS Computational Biology 4 https://doi.org/10.1371/journal.pcbi.1009823
- Reimer, L.C., Förstner, K.U. & Overmann, J. Besser forschen durch offene und FAIRe Daten. Biospektrum 28, 223 (2022). https://doi.org/10.1007/s12268-022-1725-6
- S Bray, ..., B Batut, H Rasche, ..., B Grüning, Planemo: a command-line toolkit for developing, deploying, and executing scientific data analyses, bioRxiv 2022, doi.org/10.1101/2022.03.13.483965
- M Gao, ..., B Grüning et al., Pluripotency factors determine gene expression repertoire at zygotic genome activation. Nat Commun13, 788 (2022). https://doi.org/10.1038/s41467-022-28434-1
- AC McGarvey, W Kopp, ..., A Akalin, ..., U Ohler, SA Lacadie, Single-cell-resolved dynamics of chromatin architecture delineate cell and regulatory states in zebrafish embryos, Cell Genomics, 2(1) Volume 2, Issue 1, 2022, 100083, ISSN 2666-979X, https://doi.org/10.1016/j.xgen.2021.100083
- Kopp, W., Akalin, A. & Ohler, U. Simultaneous dimensionality reduction and integration for single-cell ATAC-seq data using deep learning. Nat Mach Intell 4, 162–168 (2022). https://doi.org/10.1038/s42256-022-00443-1
- Y Perez-Riverol, ..., A Frericks-Zipper, M Eisenacher et al., The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences, Nucleic Acids Research, Volume 50, Issue D1, 7 January 2022, Pages D543–D552, https://doi.org/10.1093/nar/gkab1038
- J Schuhenn, ..., K Schork, ..., M Eisenacher et al., Differential interferon-x3b1 subtype induced immune signatures are associated with suppression of SARS-CoV-2 infection, Proceedings of the National Academy of Sciences, 119 (8)2022 https://www.pnas.org/doi/abs/10.1073/pnas.2111600119
- Holdmann J, ..., Schork K, Turewicz M, Eisenacher M et al., MicroRNAs from urinary exosomes as alternative biomarkers in the differentiation of benign and malignant prostate diseases. J Circ Biomark. 2022 Feb 10;11:5-13. https://doi.org/0.33393/jcb.2022.2317
- Höllerhage M, Stepath M, Kohl M, ..., Eisenacher M et al. Transcriptome and Proteome Analysis in LUHMES Cells Overexpressing Alpha-Synuclein, Frontiers in Neurology VOL.13 2022 https://doi.org/10.3389/fneur.2022.787059
- B Velten, ..., J Wirbel, D Bredikhin, G Zeller & O Stegle, Identifying temporal and spatial patterns of variation from multimodal data using MEFISTO.Nat Methods 19, 179–186 (2022). https://doi.org/10.1038/s41592-021-01343-9
- Koblitz J, Dirks WG, Eberth S et al. DSMZCellDive: Diving into high-throughput cell line data. F1000Research2022, https://doi.org/10.12688/f1000research.111175.1)
- Fromentin, S., … Renato Alves, … Peer Bork et al. Microbiome and metabolome features of the cardiometabolic disease spectrum. Nat Med 28, 303–314 (2022). https://doi.org/10.1038/s41591-022-01688-4
- Beier S, Fiebig A, Pommier C et al. Recommendations for the formatting of Variant Call Format (VCF) files to make plant genotyping data FAIR F1000Research 2022, 11(ELIXIR):23. https://doi.org/10.12688/f1000research.109080.1
- Martens M, Stierum R, ... Neumann S et al. ELIXIR and Toxicology: a community in development ]. F1000Research 2021, 10(ELIXIR):1129, https://doi.org/10.12688/f1000research.74502.1
- Röckel F, ..., Scholz U, ...,Lange M et al. PhenoApp: A mobile tool for plant phenotyping to record field and greenhouse observations . F1000Research 2022, 11:12 https://doi.org/10.12688/f1000research.74239.1
- M. Fahrner, MC Föll, BA Grüning, et al., Democratizing data-independent acquisition proteomics analysis on public cloud infrastructures via the Galaxy framework, GigaScience, Volume 11, 2022, giac005, https://doi.org/10.1093/gigascience/giac005
- LP Coelho, R Alves, ..., J Huerta-Cepas & Peer Bork. Towards the biogeography of prokaryotic genes. Nature 601, 252–256 (2022). doi:https://doi.org/10.1038/s41586-021-04233-4
Also of interest
- European Commission, Directorate-General for Research and Innovation, Maxwell, L., Maximising investments in health research: FAIR data for a coordinated COVID-19 response workshop report, 2022, https://data.europa.eu/doi/10.2777/726950
|
 Upcoming Events Upcoming Events
|
|
The next de.NBI training events are announced on www.denbi.de/training.
|
Impressum
|
|
Responsible for Contents
A. Puehler, A. Tauch, T. Dammann-Kalinowski
de.NBI - German Network for Bioinformatics Infrastructure c/o Forschungszentrum Juelich
Forschungszentrum Juelich - IBG-5 Branch Office at Bielefeld University, 33594 Bielefeld, Germany
Phone: +49-(0)521-106-8750
Fax: +49-(0)521-106-89046
Email:This email address is being protected from spambots. You need JavaScript enabled to view it.
Subscription
If you are interested in receiving regular updates on de.NBI - German Network of Bioinformatics Infrastructure, please subscribe to the de.NBI Quarterly Newsletter here. The mailing list will exclusively be used to distribute the de.NBI Quarterly Newsletter. To unsubscribe from the de.NBI Quarterly Newsletter, please use the unsubscribe link.
The de.NBI Quarterly Newsletter is a service of de.NBI - German Network for Bioinformatics Infrastructure for members, partners and interested public. All photos are copyright of the de.NBI administration office unless marked otherwise.
|



 We now extend a warm welcome to Michael R. Crusoe, who started his position as ELIXIR-DE Officer on April 1, 2022. Michael is a one of the Common Workflow Language co-founders and the CWL project lead. He is also a Software Carpentry instructor and Debian software packager. In addition to his employment at ELIXIR-DE, Michael is a member of the Dutch ELIXIR Node where he actively contributes to the ELIXIR Interoperability & Compute Platforms. He is also a part-time PhD student at VU Amsterdam on the topic of workflow standards in practice. Originally from Phoenix, Arizona, USA; Michael is happy to call Berlin, Germany his home after previously living in Lithuania, Romania, and Belgium.
We now extend a warm welcome to Michael R. Crusoe, who started his position as ELIXIR-DE Officer on April 1, 2022. Michael is a one of the Common Workflow Language co-founders and the CWL project lead. He is also a Software Carpentry instructor and Debian software packager. In addition to his employment at ELIXIR-DE, Michael is a member of the Dutch ELIXIR Node where he actively contributes to the ELIXIR Interoperability & Compute Platforms. He is also a part-time PhD student at VU Amsterdam on the topic of workflow standards in practice. Originally from Phoenix, Arizona, USA; Michael is happy to call Berlin, Germany his home after previously living in Lithuania, Romania, and Belgium. Sebastian Jaenicke took over his new position as Deputy Technical Coordinator in ELIXIR-DE. He succeeds Marius Dieckmann in this function, who is now employed as an NFDI software developer and cloud architect at Giessen University. Sebastian is member of the Bielefeld-Giessen (BiGi) Bioinformatics Resource Center and brings in a lot of experience field of microbial bioinformatics. His research is focusing on the design and further development of a flexible and extensible client/server-framework for the management and analysis of metagenomic datasets, also known as MGX (
Sebastian Jaenicke took over his new position as Deputy Technical Coordinator in ELIXIR-DE. He succeeds Marius Dieckmann in this function, who is now employed as an NFDI software developer and cloud architect at Giessen University. Sebastian is member of the Bielefeld-Giessen (BiGi) Bioinformatics Resource Center and brings in a lot of experience field of microbial bioinformatics. His research is focusing on the design and further development of a flexible and extensible client/server-framework for the management and analysis of metagenomic datasets, also known as MGX ( Nils-Christian Lübke from the de.NBI administration office was appointed by the CCU to represent ELIXIR-Germany’s industry liaison efforts as the new “ELIXIR-Germany Industry Officer”. Since 2022, Nils took over the role of the de.NBI Industrial Forum Coordinator from Manuel Wittchen. The de.NBI Industrial Forum provides networking opportunities at the interface between Academia and Industry. His responsibilities in this position already covered connectivity towards ELIXIR.
Nils-Christian Lübke from the de.NBI administration office was appointed by the CCU to represent ELIXIR-Germany’s industry liaison efforts as the new “ELIXIR-Germany Industry Officer”. Since 2022, Nils took over the role of the de.NBI Industrial Forum Coordinator from Manuel Wittchen. The de.NBI Industrial Forum provides networking opportunities at the interface between Academia and Industry. His responsibilities in this position already covered connectivity towards ELIXIR.