
News

The latest news regarding the de.NBI network and related activities: upcoming talks, courses, articles, job offers etc. are listed here.

- 2025-12-17
 de.NBI & ELIXIR Germany and GHGA (The German Human Genome Phenome Archive) are inviting you to the next episode of our joint Webinar Series: Galaxy for analysis of human health data.
de.NBI & ELIXIR Germany and GHGA (The German Human Genome Phenome Archive) are inviting you to the next episode of our joint Webinar Series: Galaxy for analysis of human health data.
This webinar will highlight several new features and tools added to Galaxy over the last year that further increase the platform's usability, in particular for multi-omics human health data.
Specific topics include, among a few others:
- integration of Flexynesis for deep-learning-based exploration of multi-omics data, including cBioportal studies, as a tool in Galaxy
- the new CELLxGENE integration into Galaxy
- new data resources directly accessible from the Galaxy user interface, like Tabula Sapiens reference atlas data
- enhancements around Galaxy's storage options that allow users to bring their own cloud storage and connect it to their Galaxy user account
The webinar will be held by Wolfgang Maier from the University of Freibung. The event is in English, it takes approx. 60 minutes and will be held on 14th January 2026 at 13:00 CET (available afterwards on demand). Attendance is free, but we ask for registration.

- 2025-12-15
The new de.NBI Quarterly Newsletter, has been published. It reports on main de.NBI and ELIXIR-DE activities since September 2025.

- 2025-10-21

This webinar series is a joint venture of GHGA and de.NBI & ELIXIR-DE
DNA methylation plays an important role in gene regulation. Understanding and exploring these epigenetic modifications is crucial for uncovering underlying mechanisms and gaining new insights into many diseases and disorders.
This webinar introduces approaches for analysing methylation data from common platforms such as Illumina arrays and Oxford Nanopore sequencing (ONT). We will cover practical first steps, including data processing, quality control, data visualization, as well as methods and tools for Epigenome-Wide Studies (EWAS) and episignature analyses using R.
Through real-world case studies, we will aim to illustrate and deepen the understanding of methylation analysis in practice.
This webinar is aimed at students, early career researchers, clinicians, bioinformaticians and everyone interested in methylation analysis.
The webinar will be held by Nazanin Mirza-Schreiber from the Institute of Neurogenomics (Helmholtz Zentrum München).
The event is in English, it takes approx. 60 minutes and will be held on the 10th December at 13:00 CET (available afterwards on demand). Attendance is free, but we ask for registration.

- 2025-12-09
We are pleased to announce two new job openings at DKFZ & GHGA, and kindly ask you to help spread the word:
Technical Data Steward / Bioinformatician (m/f/d, E13 TV-L, 100%)
Location: University of Tübingen
Data Steward / Bioinformatiker:in (m/w/d)
Location: DKFZ Heidelberg
If you are interested or know someone who might be, please share these opportunities!
Thank you for your support!

- 2025-11-24
Research associate computational metabolomics (m/f/d)
Salary group E13 TVL, full-time 100 % (part-time is possible), initially limited until 31.12.2027
to be filled immediately.
Research topic:
As a (bio-) informatician, you will join our research group Computational Plant Biochemistry, which focuses on Computational Metabolomics and develops methods for processing and interpreting mass spectrometry data in the context of plant biochemistry and biodiversity research.
Your tasks:
- Development of methods for the characterization and identification of metabolites
- Adaptation and use of statistical methods to detect patterns and relevant connections in metabolomics data
- Activities and Training in the framework of de.NBI and ELIXIR
Your profile:
- You hold a university PhD in bioinformatics, computer science or a related discipline
- experience in algorithms, software engineering and statistics
- You are able to program in the statistics framework R, and have worked in Python and Java
- Knowledge of metabolomics or chemistry will be an advantage
Our benefits:
- Excellent working conditions in an international environment
- Flexible and family-friendly working hours and possibility of home-office
- Offer of professional training courses and measures for skill improvement
- Compensation according to TV-L (including annual special payment)
- Contribution to your company pension plan (VBL)
- On-site opportunities for health promotion
How to apply and information:
For further information, please contact Dr. Steffen Neumann , Tel.: +49 (0) 345 5582-1770, e-mail:
Please, submit a single pdf document (letter of motivation, CV, certificates, publication list, etc.) quoting reference number 21/2025 until December 22nd, 2025 to
Foreign qualifications must comply with German standards (TV-L-EntgeltO Protokollerklärung Nr. 1 Absatz 4) and be certified (equivalence test in Germany, subject to a fee) and presented to IPB Human Resources at the time of hiring: (https://www.kmk.org/zab/central-office-for-foreign-education).
Who we are:
The Leibniz Institute of Plant Biochemistry (IPB) is a non-university research institution of the Leibniz Association on the Weinberg Campus of the Martin Luther University Halle-Wittenberg. As a foundation under public law, the IPB is under direct supervision of the state of Saxony-Anhalt.
The IPB is an internationally recognized research institution and consists of four scientific Departments, additional independent junior research groups, and the Department Administration & Infrastructure (about 200 employees, including about 40 PhD students). Research at the IPB aims to understand the (bio)chemical basis of plant resilience and performance in challenging environments related to climate change. The IPB offers excellent research facilities and state-of-the-art infrastructure to investigate the chemical diversity, biochemical interactions, and biological roles of small natural molecules in plants and fungi, with an emphasis on specialized metabolites, chemical mediators, and relevant molecular networks of functional gene and protein regulation (https://www.ipb-halle.de/en/).
Diversity, family and equal opportunities:
The IPB aspires to the goal of equal opportunity, diversity, and the promotion of work-life balance as it was awarded the certificate "Total E-Quality". In addition, the IPB is a member of the nationwide company network "Success Factor Family" and of the “Diversity Charter” (Charta der Vielfalt).
Applications from disabled persons or applicants with equivalent status are expressly welcome.
Further information can be found at: https://www.ipb-halle.de/en/institute/
Data protection:
Please note, the data protection information for applicants (m/f/d) according to Article 13 and 14 GDPR on data protection processing during the application process:
https://www.ipb-halle.de/en/career/data-protection-information-for-applicants/
Leibniz-Institut für Pflanzenbiochemie
Weinberg 3
06120 Halle

- 2025-10-21

This webinar series is a joint venture of GHGA and de.NBI & ELIXIR-DE
This webinar is intended for researchers who are new to the field of spatial transcriptomics. It provides an introduction to commonly used image-based spatial omics technologies and demonstrates the data processing workflow through a practical example. Topics include steps such as gene panel design, cell segmentation, doublet detection, and others.
Given the complexity of spatial transcriptomics, the webinar will focus primarily on data preprocessing steps and will only briefly touch upon downstream analyses such as domain detection.
The webinar will be held by GHGA workflows expert Florian Heyl. The event is in English, it takes approx. 60 minutes and will be held on the 20th November 2025 at 13:00 CET (available afterwards on demand). Attendance is free, but we ask for registration.

- 2025-10-29
A doctoral and a postdoctoral position in computational biology / biomathematics / biosystems engineering are available in Steffen Klamt’s group at the Max Planck Institute for Dynamics of Complex Technical Systems in Magdeburg (Germany). Our group develops computational methods and tools for analyzing and redesigning the microbial metabolism, with applications in metabolic engineering and bioprocess optimization.
Your tasks
For two interdisciplinary research projects in the field of computer-aided metabolic engineering, we are seeking a talented doctoral student and a postdoctoral researcher. Your responsibilities will include:
- Building stoichiometric / kinetic metabolic models of selected microorganisms.
- Applying various computational methods to analyze these models and identify suitable intervention strategies maximizing the synthesis of specific products.
- Using experimental data to improve the models and to support an iterative design-build-test-learn cycle.
- Integrating concepts from bioprocess optimization with metabolic engineering strategies.
Your qualification
The candidates should have an excellent degree or/and experience in at least one of the following fields:
- Computational biology
- Systems biology
- Biomathematics
- Biosystems and metabolic engineering
- Bioprocess engineering
Experience in the following areas is particularly advantageous:
- Mathematical and kinetic modeling of metabolic networks
- Mathematical optimization (linear and mixed-integer (non-)linear programming)
- Programming experience in MATLAB and/or Python
- Model-driven strain design and metabolic engineering
- Bioprocess and biosystems engineering
Our offer
- Fixed-term employment contract in the public sector (German TVöD E13)
- Working in a creative and inspiring atmosphere
- Opportunities for networking with international partners
- Research with state-of-the-art equipment
- Compatibility of family, career and caring for relatives through flexible working hours and appropriate conditions
- Health-promoting and health-maintaining measures as part of our occupational health management
Your application
Interested applicants are encouraged to submit their applications as soon as possible. However, there is no strict deadline, the call will remain open until the position is filled. Applications should be submitted as a single PDF file, including
- An application letter
- Curriculum vitae (CV)
- Academic certificates
- A brief description of scientific achievements (if applicable)
- Contact information for two references (if possible).
Applications can be submitted at https://jobs.mpi-magdeburg.mpg.de/zdn86
More information about our group and research can be found on the following website:
http://www.mpi-magdeburg.mpg.de/arb.
Contact person for further information: Dr. Ing. Steffen Klamt (+49-391-6110-480).

- 2025-09-30

In this webinar, you can learn more about how the German Network for Bioinformatics Infrastructure (de.NBI) and ELIXIR Germany are at the forefront of advancing bioinformatics and creating a robust Research Data Management (RDM) ecosystem in the life sciences. As part of a European ELIXIR infrastructure, they provide a comprehensive portfolio of services, tools, training opportunities, and platforms supporting researchers in managing, analyzing, and sharing their data efficiently and securely.
The de.NBI Cloud works hand in hand with the recently established de.KCD (Competence Center for Cloud Technologies and Data Management). This center provides expertise, training, and best practices in cloud computing and RDM. Together, they deliver scalable computing and storage resources that meet the growing demands of big data, reproducible research, and interdisciplinary collaboration.
This presentation offers a practical introduction to getting started with the de.NBI Cloud, including step-by-step guidance on registration, project proposal submission, and gaining access to resources. The SimpleVM self-service platform of the de.NBI Cloud enables researchers - even with minimal technical background - to easily launch, configure, and manage virtual machines tailored to their computational projects.
Looking ahead, the webinar will highlight how the de.NBI cloud intends to drive forward the integration of the de.NBI Cloud into RDM frameworks, with a strong focus on implementing the FAIR data principles. Future tasks will include best practices for organizing project data, the use of cloud-based workflows, and secure data exchange between institutions. In addition, we plan to expand our focus on the responsible handling of sensitive data in order to keep pace with advancing data protection and compliance requirements.
This webinar is aimed at students, early career researchers, clinicians, bioinformaticians and everyone interested in using the de.NBI Cloud.
The webinar will be held by Daniel Wibberg from Forschungszentrum Jülich (de.NBI & ELIXIR-DE) .
The event is in English, it takes approx. 60 minutes and will be held on 14th October 2025, 13:00 CET (available afterwards on demand). Attendance is free, but we ask for registration.

- 2025-09-19
 We're excited to announce a new webinar series designed to help researchers navigate the challenges of modern life science data. This series is a collaborative effort between the German Human Genome-Phenome Archive (GHGA) and de.NBI & ELIXIR Germany.
We're excited to announce a new webinar series designed to help researchers navigate the challenges of modern life science data. This series is a collaborative effort between the German Human Genome-Phenome Archive (GHGA) and de.NBI & ELIXIR Germany.
Today's life science research produces a massive amount of complex data. To make the most of it, researchers need to use strong bioinformatics methods and smart data management. This series will provide the practical guidance, tools, and best practices you need to do just that.
What to Expect
Each webinar will feature experts invited by GHGA, de.NBI & ELIXIR Germany who will share their knowledge and offer different perspectives on secure data handling and effective bioinformatics workflows.
The series is perfect for early career researchers in bioinformatics and biological data management, but is open to anyone who wants to dive deeper into the field.
Series details
Topics: The webinars will cover a range of topics, including:
- Best practices and new trends in bioinformatics
- Effective strategies for biological data management
- Case studies of successful bioinformatics applications
Format: Each session will be around 60 minutes, including a 35-minute talk and a 20-minute Q&A session with the experts.
When: We will host 8-10 webinars per year, with one session every four to six weeks.
Where: All webinars will be given as an interactive video-conference, the free registration link will be posted on https://www.ghga.de/events and https://www.denbi.de/training
Recordings: Edited video recordings will be available on the YouTube channels of both GHGA (@ghga-thegermanhumangenome) and de.NBI & ELIXIR Germany (@denbi5170).
Dates 2025
14th October 25 at 13:00 CEST “Building a RDM Ecosystem: The Role of de.NBI & ELIXIR Germany and how to use the de.NBI Cloud” with Daniel Wibberg
20th November 25 at 13:00 CET “Data processing and beyond for image-based spatial transcriptomics technologies” with Florian Heyl
10th December 25 at 13:00 CET “Methylation Analysis: A Practical Guide for Beginners” with Nazanin Mirza-Schreiber
Stay tuned for more information on our first webinar and how to register!
- 2025-09-04
The new de.NBI Quarterly Newsletter, has been published. It reports on main de.NBI and ELIXIR-DE activities since July 2025.
- 2025-06-16
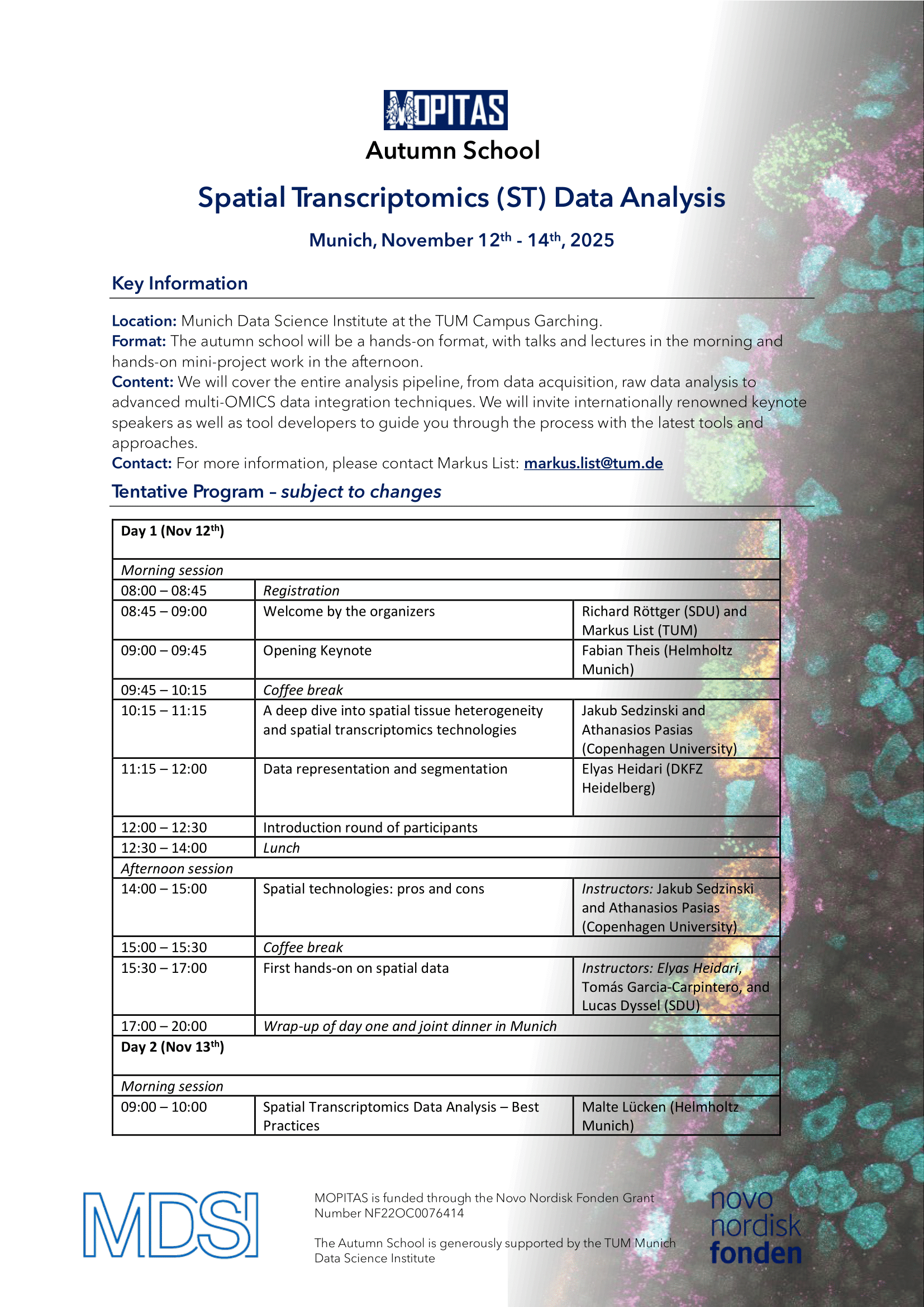
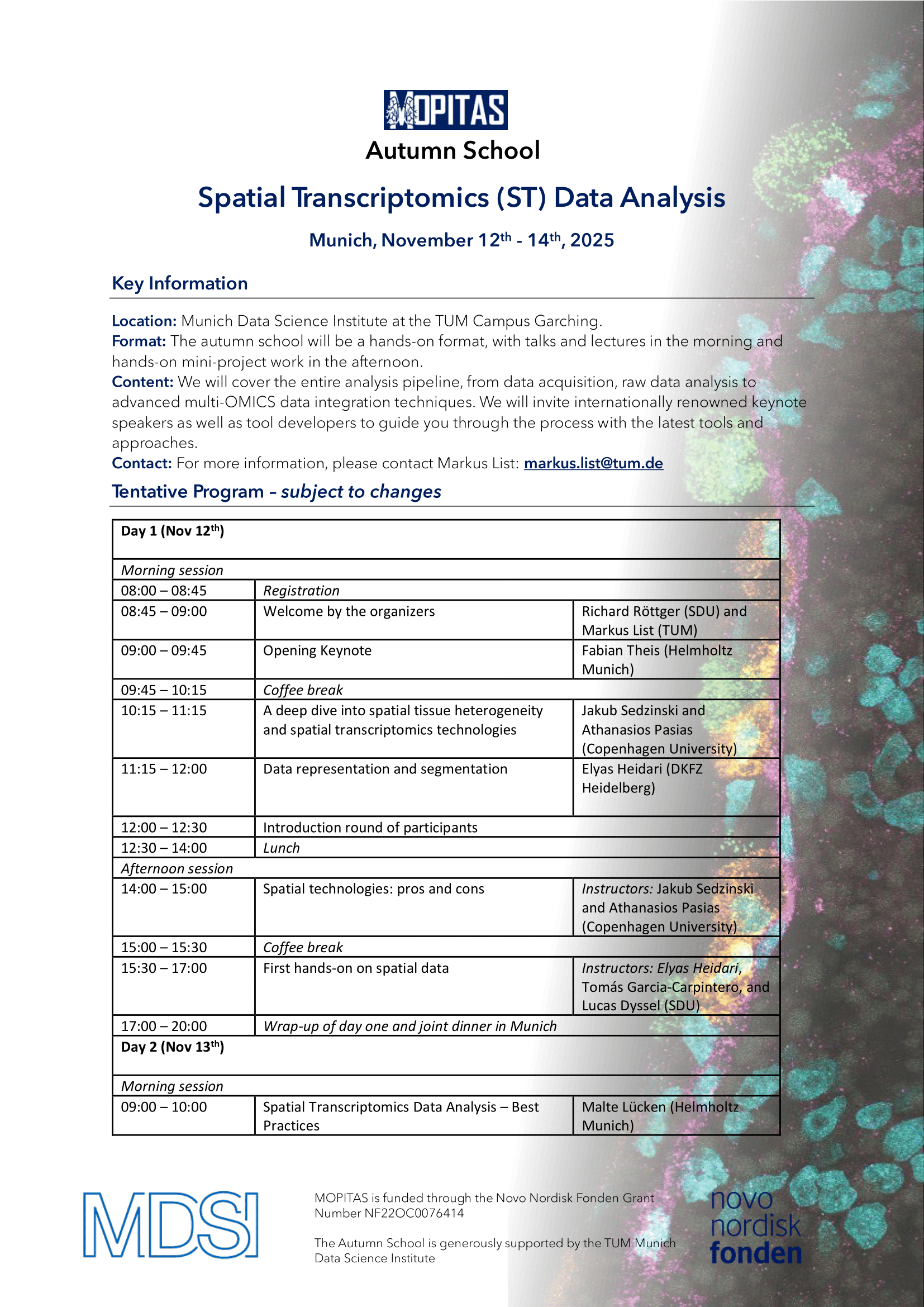
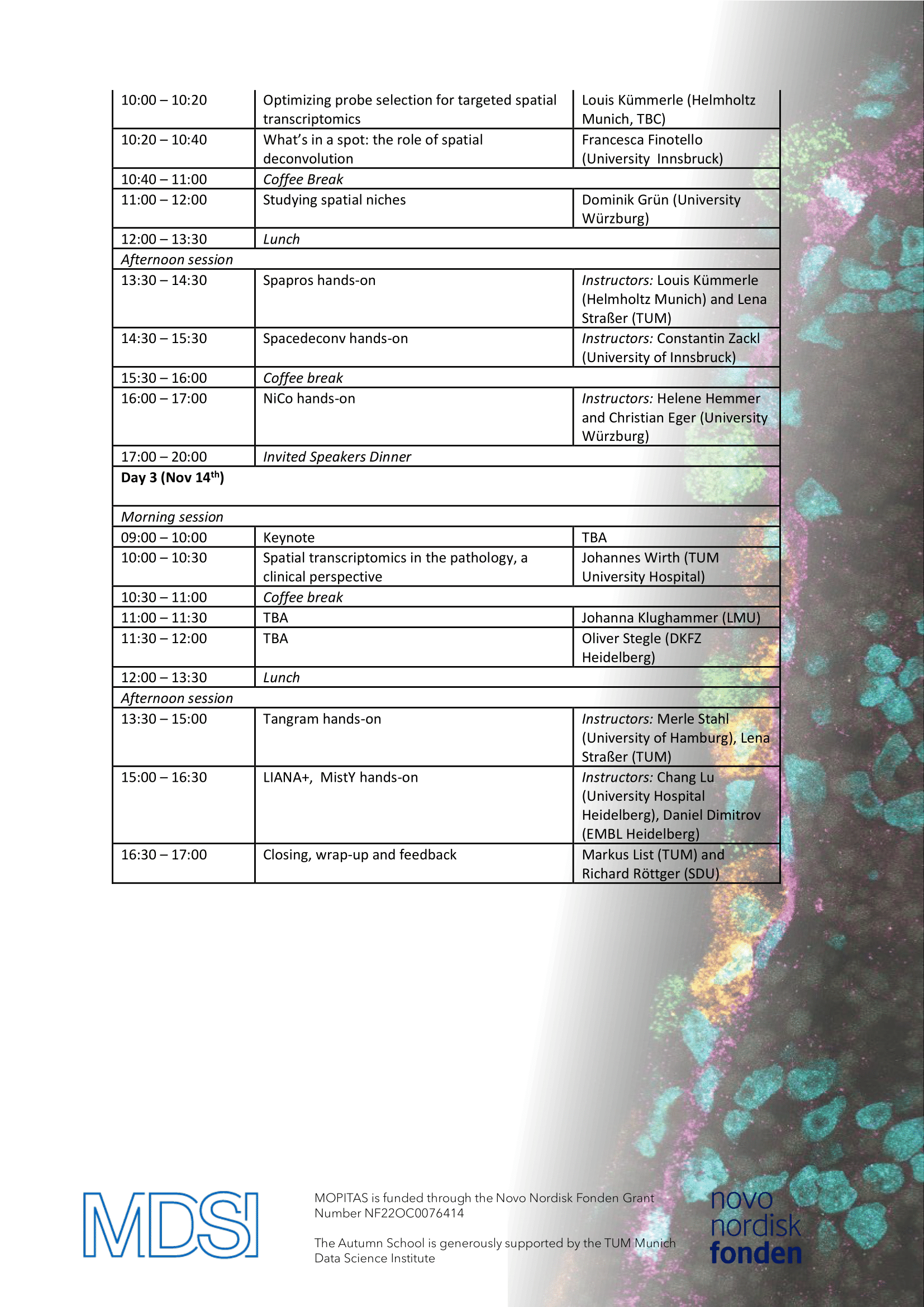
Location: Munich Data Science Institute, TUM Campus Garching
Date: November 12-14, 2025
We are pleased to invite you to our upcoming Autumn School!This immersive event is designed for students, researchers, and professionals eager to deepen their expertise in data science and multi-OMICS analysis.
Format:
The Autumn School will feature a dynamic, hands-on format. Mornings will be dedicated to insightful talks and lectures, while afternoons will focus on collaborative mini-projects, allowing participants to apply what they’ve learned in a practical setting.
Whether you're looking to expand your technical skills or connect with experts and peers in the data science community, this Autumn School offers a unique opportunity to learn, collaborate and innovate. Certificates of attendance will be issued after the completion of the Autumn School. Registration is closing on July 1st, 2025: https://event.sdu.dk/mopitas-school/conference
We look forward to welcoming you!
Prof. Markus List (TUM, DE) & Prof. Richard Röttger (SDU, DK)

- 2025-06-13
The new de.NBI Quarterly Newsletter, has been published. It reports on main de.NBI and ELIXIR-DE activities since March 2025.

- 2025-06-12

| 2025-09-22 | Leveraging Cloud Computing for Bioinformatics: A SimpleVM Workshop Featuring a Metagenomics Use Case - GCB2025 | Düsseldorf |
| 2025-09-22 | ProteinsPlus – Supporting Structure-Based Design on the Web - GCB 2025 | Düsseldorf |
| 2025-09-22 | Computational Pangenomics Workshop - GCB 2025 | Düsseldorf |

- 2025-05-15
An der Technischen Hochschule Mittelhessen ist am Campus Gießen im Fachbereich Mathematik, Naturwissenschaften und Informatik (MNI) folgende Stelle
W2-Professur
mit folgendem Fachgebiet
Informatik mit Schwerpunkt Bioinformatik
Ref. Nr. B25-006
baldmöglichst zu besetzen.
Die Technische Hochschule Mittelhessen gehört zu den größten Hochschulen angewandter Wissenschaften in Deutschland. Zur Verstärkung unseres Kollegiums suchen wir eine Persönlichkeit mit fundierter Praxiserfahrung in den Bereichen der Bioinformatik und Data Science. Sie/Er hat das Berufungsgebiet in Forschung und Lehre (auch in den Grundlagenveranstaltungen) zu vertreten und dabei dessen interdisziplinärem Charakter Rechnung zu tragen.
Erwartet werden fundierte Kenntnisse in mindestens zwei der folgenden Gebiete:
· Maschinelles Lernen / Deep Learning
· Molekularbiologische Datenanalyse / *Omics
· Systembiologie / Molecular Modeling
· Data Science im Bereich der Life Sciences.
Außerdem wird die Fähigkeit und Bereitschaft erwartet, grundständige Lehrveranstaltungen in den Bachelor- und Masterstudiengängen der Bioinformatik, Informatik und Data Science durchzuführen.
Als Technische Hochschule steht der Praxisbezug im Fokus der Ausbildung der Studierenden, so dass erfolgreiche einschlägige praktische Tätigkeiten in der Industrie bzw. in der industrienahen Forschung, beispielsweise in der Führung von wissenschaftlichen Arbeitsgruppen, unabdingbar sind.
Wir erwarten von unseren Professorinnen und Professoren:
- Engagement und Initiative durch hohe Präsenz an der Hochschule und intensive Betreuung der Studierenden, damit verbunden die Bereitschaft zur Wohnsitznahme im Raum Gießen
- Kontinuierliche Weiterbildung in Fachwissenschaft und Hochschuldidaktik
- Beteiligung an internationalen Aktivitäten der Hochschule
- Übernahme von Verantwortung im Rahmen der akademischen Selbstverwaltung
- Befähigung und Neigung zur Forschung und Einwerbung von Drittmitteln
- Lehrveranstaltungen an allen Hochschulstudienorten (bei entsprechendem Bedarf)
- hohe Präsenz der Lehrenden an der Hochschule
- Gute Deutsch- und Englischkenntnisse, die eine qualifizierte Lehre in deutscher und englischer Sprache ermöglichen, werden vorausgesetzt.
Wir bieten unseren Professorinnen und Professoren:
- Einarbeitung durch Teilnahme an hochschuldidaktischer Grundschulung
- Leistungsorientiertes Entgelt
- Arbeiten in angenehmer und kollegialer Atmosphäre
- Mitarbeit in den Kompetenzzentren mit fächerübergreifender praxisbezogener Forschung
- Möglichkeit zu Tätigkeiten in der Weiterbildung
Es gelten die folgenden formalen Einstellungsvoraussetzungen gemäß § 68 Hessisches Hochschulgesetz (HessHG):
- Abgeschlossenes Hochschulstudium des entsprechenden Wissenschaftsgebietes
- Pädagogische Eignung, die in der Regel durch Erfahrungen in der Lehre, hochschuldidaktische Qualifikationen und durch eine Probeveranstaltung (Probevortrag, Probelehrveranstaltung) nachgewiesen wird
- Besondere Befähigung zu wissenschaftlicher Arbeit, die in der Regel durch die Qualität der Promotion nachgewiesen wird
- besondere Leistungen bei der Anwendung oder Entwicklung wissenschaftlicher Erkenntnisse und Methoden in einer mindestens fünfjährigen einschlägigen beruflichen Praxis, von der mindestens drei Jahre außerhalb des Hochschulbereichs ausgeübt worden sein müssen oder
- zusätzliche wissenschaftliche Leistungen
Die Stelle steht unbefristet zur Verfügung. Bei der ersten Berufung auf eine Professur erfolgt die Einstellung zunächst in einem Beamtenverhältnis auf Probe bzw. in einem Angestelltenverhältnis. Im Übrigen wird auf § 67 Hessisches Hochschulgesetz Bezug genommen.
Die Technische Hochschule Mittelhessen strebt im Bereich des wissenschaftlichen Personals die Erhöhung des Anteils der Frauen an. Entsprechend qualifizierte Wissenschaftlerinnen werden nachdrücklich um ihre Bewerbung gebeten.
Schwerbehinderte Bewerberinnen und Bewerber werden bei gleicher Eignung besonders berücksichtigt.
Als familienfreundliche Hochschule unterstützen wir unsere Beschäftigten bei der Vereinbarkeit von Familie und Beruf. Eine Beschäftigung mit reduzierter Arbeitszeit ist im Rahmen der gesetzlichen Bestimmungen möglich.
Damit wir uns ein umfassendes Bild von Ihnen machen können, sollte Ihre Bewerbung folgende Unterlagen beinhalten: Anschreiben, tabellarischer Lebenslauf, Zeugniskopien, Beurteilungen, lückenlose Tätigkeitsnachweise, Veröffentlichungen sowie ein Lehr- und Forschungskonzept. Bei einem im Ausland erworbenen Hochschulabschluss, bitten wir Sie, Ihrer Bewerbung eine Zeugnisbewertung beizufügen.
Ihre aussagekräftige Bewerbung richten Sie bitte ausschließlich über unser Bewerbermanagementsystem bis 28.05.2025 unter Angabe der Referenznummer an den Präsidenten der Technischen Hochschule Mittelhessen.
Die Vereinbarkeit von Familie und Beruf ist uns ein Anliegen. Im Rahmen des Audits "Familiengerechte Hochschule" arbeiten wir an der Weiterentwicklung entsprechender Strukturen.
Mit dem Absenden einer Bewerbung willigen Sie ein, dass Ihre Daten zum Zwecke des Stellenbesetzungsverfahrens gespeichert und verarbeitet werden. Bei Fragen zur Stelle wenden Sie sich bitte an Frau Wagner, 0641 309-1061. Online-Bewerbung

- 2025-03-13
The new de.NBI Quarterly Newsletter, has been published. It reports on main de.NBI and ELIXIR-DE activities since December 2024.

- 2025-02-17
The Computational Plant Biochemistry research group at the IPB is seeking applications to fill the position as
Research associate (computational metabolomics, m/f/d)
PostDocs: Salary group E13 TVL, full-time 100 %, limited for two years
PhD students: Salary group E13 TVL, 75 %, limited for three years
to be filled immediately.
Research topic:
As a (bio-) informatician, you will join our research group Computational Plant Biochemistry, which focuses on Computational Metabolomics and develops methods for processing and interpreting mass spectrometry data in the context of plant biochemistry research.
Your tasks:
- Development of methods for the characterization and identification of metabolites
- Adaptation and use of statistical methods to detect patterns and relevant connections in metabolomics data
- Integration of data from multiple Omics experiments
Your profile:
- You hold a university degree (master or PhD) in bioinformatics or computer science, with experience in algorithm and software engineering and machine learning
- You are able to program in the statistics framework R, and have worked in Python and Java
- Knowledge in metabolomics or chemistry will be an advantage
Our benefits:
- Excellent working conditions in an international environment
- Flexible and family-friendly working hours and possibility of home-office
- Offer of professional training courses and measures for skill improvement
- Compensation according to TV-L (including annual special payment)
- Contribution to your company pension plan (VBL)
- On-site opportunities for health promotion
Who we are:
The Leibniz Institute of Plant Biochemistry (IPB) is a non-university research institution of the Leibniz Association on the Weinberg Campus of the Martin Luther University Halle-Wittenberg. As a foundation under public law, the IPB is under direct supervision of the state of Saxony-Anhalt.
The IPB is an internationally recognized research institution and consists of four scientific Departments, additional independent junior research groups, and the Department Administration & Infrastructure (about 200 employees, including about 40 PhD students). Research at the IPB aims to understand the (bio)chemical basis of plant resilience and performance in challenging environments related to climate change. The IPB offers excellent research facilities and state-of-the-art infrastructure to investigate the chemical diversity, biochemical interactions, and biological roles of small natural molecules in plants and fungi, with an emphasis on specialized metabolites, chemical mediators, and relevant molecular networks of functional gene and protein regulation (https://www.ipb-halle.de/en/).
How to apply and information:
For further information, please contact Dr. Steffen Neumann , Tel.: +49 (0) 345 5582-1770, e-mail:
Please, submit a single pdf document (letter of motivation, CV, certificates, publication list, etc.) quoting reference number 05/2025 until March 31st, 2025 to
Foreign qualifications must comply with German standards (TV-L-EntgeltO Protokollerklärung Nr. 1 Absatz 4) and be certified (equivalence test in Germany, subject to a fee) and presented to IPB Human Resources at the time of hiring: (https://www.kmk.org/zab/central-office-for-foreign-education).
Diversity, family and equal opportunities:
The IPB aspires to the goal of equal opportunity, diversity, and the promotion of work-life balance as it was awarded the certificate "Total E-Quality". In addition, the IPB is a member of the nationwide company network "Success Factor Family" and of the “Diversity Charter” (Charta der Vielfalt).
Applications from disabled persons or applicants with equivalent status are expressly welcome.
Further information can be found at: https://www.ipb-halle.de/en/institute/
Data protection:
Please note, the data protection information for applicants (m/f/d) according to Article 13 and 14 GDPR on data protection processing during the application process:
https://www.ipb-halle.de/en/career/data-protection-information-for-applicants/

- 2025-01-08
Happy New Year! We are back from the Christmas break and ready to make 2025 amazing! Let's get started!
Follow us now also on Bluesky (@denbi.bsky.social) and Mastodon (@deNBI). #BackToWork #2025

- 2024-06-19
The new de.NBI Quarterly Newsletter, has been published. It reports on main de.NBI and ELIXIR-DE activities since September 2024.
Main topics:
- Review de.NBI All Hands Meeting 2024
- The German Competence Center Cloud Technologies for Data Management and Processing (de.KCD)
- New ELIXIR Germany TrC and TeC deputies
- ...and much more!
- 2024-11-14
An international research team led by the de.NBI member IPK Leibniz Institute Gatersleben has published a comprehensive pangenome of barley, featuring long-read sequence assemblies of 76 wild and domesticated genomes along with short-read data from 1,315 genotypes. This expanded catalogue highlights structural variants, such as gene copy number variations, which control traits like disease resistance and starch mobilization. Published in "Nature," the study sheds light on barley's evolution and breeding potential, emphasizing how genetic diversity can arise post-domestication to meet agricultural demands. As one of the top global crops, barley's structural diversity is crucial for adapting to harsh environments, offering insights into future crop improvement under changing climates. This project, coordinated by IPK and involving 80 scientists from 12 countries, underlines the institute's leadership in crop genomics. The research also benefited from tools and resources provided by de.NBI - CATS and e!DAL-PGP - enhancing the analysis of complex genomic data.

- 2024-10-24
Educators:
de.NBI Cloud group
Date:
2 - 6 December 2024
Location:
Online
Contents:
For more than 6 years, the de.NBI Cloud has provided free of charge computing resources to the life sciences community in Germany. Scientists involved in various research topics, such as metagenomics, medical imaging, epigenetics, and plant biology benefit from the opportunities provided by the de.NBI Cloud.
With our annual user meeting, we would like to highlight topics of interest to our user community.
This year we are focusing exclusively on experienced users, while new users will be offered recurring introductory courses in SimpleVM and OpenStack starting next year. We will give our current users the opportunity to deepen their understanding of best practices in cloud computing. There will be workshops on advanced topics such as container orchestration with Kubernetes or handling cloud-based clusters with BiBiGrid.
The de.NBI Cloud User Meeting supports the interaction with our growing community. Thus, we would like to hear from you about the specific needs of the scientific life sciences community, allowing us to shape the future of the de.NBI Cloud according to sophisticated and specialized use cases.
This year, we are organizing the de.NBI Cloud User Meeting as an online event via zoom in December from 02.12 to 06.12.
The scope of this meeting includes advanced workshops tailored for experienced users and opportunities to engage with experts on diverse cloud computing topics
We look forward to welcoming experienced researchers from many different bioinformatics and life-science research areas to delve deeper into cloud computing and the de.NBI Cloud.
Learning goals
• Working with the de.NBI cloud project types OpenStack and Kubernetes
• Working with Bioinformatics tools and workflows
Prerequisites:
• The scope of this meeting includes advanced workshops tailored for experienced users and opportunities to engage with experts on diverse cloud computing topics
• de.NBI Cloud User account (see instructions at https://cloud.denbi.de/wiki/registration/
• Laptop for local program execution, browser-based access and ssh-based access
Keywords:
de.NBI Cloud, Linux, OpenStack, Ansible, Kubernetes, BiBiGrid, Research Environments
Tools:
de.NBI Cloud
Contact:
Registration:
https://events.hifis.net/e/clum7

- 2024-06-19
The new de.NBI Quarterly Newsletter, has been published. It reports on main de.NBI and ELIXIR-DE activities since July 2024.
Main topics:
- 3rd BioHackathon Germany – Registration
- de.NBI/ELIXIR-DE All Hands Meeting 28-29 Nov 2024
- Annual Meeting of de.NBI Industrial Forum
- ...and much more!

- 2024-09-16
Our team is looking for a
PhD student in computational proteomics (f/m/d) for non-clinical safety assessment with ProteomicsDB
with a good background in bioinformatics, data science, data visualization, and big data analytics.
ProteomicsDB (https://www.proteomicsdb.org/) is an internationally well-reputed publicly available knowledgebase that provides information about proteins and other bio-molecules. It is designed to integrate data from mass spectrometry-based experiments and other omics to offer comprehensive views of proteomes, especially in the context of perturbations, like drugs. We (https://www.mls.ls.tum.de/en/compms/home/) are currently looking to fill an open position that will be tasked to extend ProteomicsDB to serve as the central database for an EU-funded project aiming to reduce and replace non-human primates in non-clinical safety assessment (https://ec.europa.eu/info/funding-tenders/opportunities/portal/screen/opportunities/topic-details/horizon-ju-ihi-2023-04-01-two-stage). In this non-competitively funded project, many EU partners from academia, biotech, pharma, and service providers combine forces to share and obtain knowledge of minipigs. The goal is to facilitate the development of innovative solutions by improving the translational understanding between minipigs versus non-human primates and humans, including further understanding of the minipig immune system, with the overall aim to replace, reduce, and refine the use of animals in non-clinical safety assessment.
The successful candidate will become a key member of a strong interdisciplinary team of bioinformatician that focus on the development and application of novel approaches in proteomic and biomedical research. This position will strengthen our ongoing development of ProteomicsDB focusing on the backend or frontend development. In light of this, the most important aspects of the job description are:
• Data import, maintenance, and integration of multi-omics data using standard data formats and ontologies
• Service engineering, infrastructure maintenance, and full stack development for ProteomicsDB
• Development of novel multi-omics integration approaches e.g. though machine and deep learning (ML/DL) for non-clinical safety assessment of drugs
• Development of novel data analysis routines e.g. through large language models (LLMs)
Requirements: Candidates must hold a master’s degree in Data Engineering, Data Science, Bioinformatics, Informatics, or a related discipline. Essential skills include theoretical knowledge of and practical skills in statistical analysis, data mining, data integration, machine learning, programming, backend or frontend development, and database design. Additional desirable skills include a sound understanding of proteomic or related technologies as well as basic biological and (bio)chemical concepts. Interest in understanding technologies e.g. proteomics and metabolomics using mass spectrometers is expected. We are looking for a self-motivated and broadly interested individual with high potential and a strong sense of responsibility. Flexibility and the ability to work in a fast-paced environment on multiple scientific and infrastructure projects are essential. Good inter-cultural and inter-personal communication skills as well as the ability to present in English are also important.
Our offer: You will join a young and highly motivated team of interdisciplinary bioinformaticians that use the latest proteomic approaches to gain insight into the biological processes that govern life. You will further be connected to the Munich Data Science Institute (MDSI) at the Technical University of Munich (TUM) to foster and facilitate exchange. You will also be connected to the SAP University Competence Center (UCC) at the TUM to assist in the hardware/software maintenance of ProteomicsDB. TUM is one of the best academic institutions in Germany and offers a stimulating work environment and excellent future perspectives. The position is available as soon as possible. Remuneration is in accordance with the TV-L E13 according to the professional qualification.
Equal opportunity: TUM is aiming to increase the proportion of women and, therefore, expressly welcomes applications from women. The position is also suitable for severely disabled persons. Severely disabled applicants will be given preference if their suitability, qualifications and professional performance are otherwise essentially equal.
Application: Applications should include a motivational statement (maximum one page), a curriculum vitae summarizing qualifications and experience, copies of degrees/university transcripts, names and email addresses of at least one referee. Applications should be sent as a single PDF to Prof. Dr. Mathias Wilhelm (
Notes on data security: When you apply for a position with the Technical University of Munich (TUM), you are submitting personal information. With regard to personal information, please take note of the Datenschutzhinweise gemäß Art. 13 Datenschutz-Grundverordnung (DSGVO) zur Erhebung und Verarbeitung von personenbezogenen Daten im Rahmen Ihrer Bewerbung. (data protection information on collecting and processing personal data contained in your application in accordance with Art. 13 of the General Data Protection Regulation (GDPR). By submitting your application, you confirm that you have acknowledged the above data protection information of TUM

- 2024-08-23
Your Job:
- Work on a broad spectrum of methods and applications of metagenomics and cloud computing
- Processing, analyzing and interpreting metagenome data from various NGS platforms (e.g. Illumina, ONT, PacBio) as part of various collaborative projects
- Development of analysis workflows with integration of container and cloud technologies (e.g. Docker, Aptainer, S3, Openstack, Ansible)
- Supporting the working group in the area of administrative project work such as writing applications and reports and participating in project meetings
- Supporting the working group in planning and organizing training courses for researchers at different career levels and from different disciplines
- Publication of scientific results in international journals and traveling to conferences
- Exchange with internal and external project partners at IBG-5 and Bielefeld University
- (Co)-supervision of doctoral students
Your Profile:
- A completed scientific university degree (Master`s degree) in the field of bioinformatics, computer science, mathematics, engineering, natural sciences or economics, or a related degree programme, followed by a completed doctorate/Phd studies
- Experience in big data analyses in connection with cloud technologies and workflows
- Good knowledge of sequence analysis and metagenomics, especially in the processing of Illumina and ONT data
- Very good programming skills (e.g. in Python or comparable) and knowledge of relevant technologies (e.g. Git, Docker, Nextflow)
- Very confident with Linux/Unix
- Fluent written and spoken English
- High degree of independence and willingness to familiarise yourself with complex topics
- Very reliable and conscientious working style
- Good publication performance
- Willingness to actively impart knowledge, e.g. by supporting the planning and implementation of training courses
- Willingness to travel and participate in conferences, networking events and project meetings
Our Offer:
We work on the very latest issues that impact our society and are offering you the chance to actively help in shaping the change! We support you in your work with:
- Highly interesting, interdisciplinary, varied work in a highly topical subject area
- Participation in project meetings and conferences
- Strong support and mentoring for building a future career in academia and/or industry
- Further development of your personal strengths, e.g. through an extensive range of training courses; a structured further training program and networking opportunities especially for PhD students via JuDocS, the Jülich centre for PhD students and supervisors: https://www.fz-juelich.de/judocs
- Optimal conditions for a good work-life balance and a family-conscious company policy
- The possibility to work flexibly (in terms of location), e.g. in a home office, is generally given after consultation and in accordance with the tasks and (on-site) appointments at hand
- Academic achievements and a company pension scheme
- Targeted services for international employees, e.g. through our International Advisory Service
In addition to exciting tasks and a collaborative working atmosphere at Jülich, we have a lot more to offer: https://go.fzj.de/benefits
We offer you an exciting and varied role in an international and interdisciplinary working environment. The position is initially for a fixed term of 2 years, with possible long-term prospects. Salary and social benefits will conform to the provisions of the Collective Agreement for the Public Service (TVöD-Bund), pay group EG 13, depending on the applicant’s qualifications and the precise nature of the tasks assigned to them.
We welcome applications from people with diverse backgrounds, e.g. in terms of age, gender, disability, sexual orientation / identity, and social, ethnic and religious origin. A diverse and inclusive working environment with equal opportunities in which everyone can realize their potential is important to us.
More information here: https://www.fz-juelich.de/en/careers/jobs/2024-278

- 2024-08-14
Reference number: 2024-0231
- Full-time
- German Human Genome-Phenome Archive (GHGA)
“Research for a life without cancer" is our mission at the German Cancer Research Center. We investigate how cancer develops, identify cancer risk factors and look for new cancer prevention strategies. We develop new methods with which tumors can be diagnosed more precisely and cancer patients can be treated more successfully. Every contribution counts – whether in research, administration or infrastructure. This is what makes our daily work so meaningful and exciting.
The German Cancer Research Center is seeking for the recently launched infrastructure initiative German Human Genome-Phenome Archive (GHGA) a
GHGA (www.ghga.de) is part of the national program for research data infrastructures (NFDI). As a node of the federated European Genome-Phenome Archive, GHGA will contribute towards the establishment of an international infrastructure for human genome data. These activities are closely coordinated with leading international initiatives and networks, such as the European Genomics Data Infrastructure (GDI), and the Global Alliance for Genomics and Health (GA4GH). This infrastructure will support genomics data hubs with software tools for secure data / metadata storage, interactive data portals with data visualization, and streamlined data deposition and acquisition solutions. GHGA is also part of genom.DE and the upcoming German national genome sequencing model project (“Modellvorhaben Genomsequenzierung"). The GHGA main office, which is coordinated by Prof. Oliver Stegle (Division of Computational Genomics and Systems Genetics), is located at DKFZ in close coordination with the European Molecular Biology Laboratory (EMBL, Prof. Jan Korbel).
YOUR TASKS
We are looking for a Data Steward / Data Manager with a bioinformatics background to operate the data management system within GHGA and the new model project genome sequencing. As part of the role, you will take responsibility to moderate day to day activities on the data portal, optimize and extend existing operation protocols, and also guide the product development by incorporating user feedback for new versions of the GHGA toolset. The successful candidate will be part of an interdisciplinary research and data management team developing and applying a diverse range of state-of-the-art methodology to implement and maintain bioinformatics workflows in a cloud compute environment. The role will involve close cooperation with other partners in the GHGA network, as well as international networks and initiatives.
Your tasks:
- Identification of suitable datasets and analysis of their potential for data sharing
- Quality control of datasets and preparation of data and metadata for submission into EGA or GHGA
- Development and implementation of SOPs for data management of human omics data as part of the GHGA infrastructure
- Strategy and processes for data ingest, metadata validation and quality control of data
- Handling of data access requests and managing the necessary administrative and legal processes
- Closely follow the field of secure data handling and sharing and interaction with international initiatives such as the Global Alliance for Genomics and Health (GA4GH)
We offer the opportunity to help shape one of the most important emerging scientific data infrastructures for the storage and exchange of omics data in Germany.
Your possibilities:
- Get exposed to a plethora of fields ranging from AI-guided clinical decision making to modern web development technologies
- Work with an interdisciplinary team of experts bringing state-of-the-art biomedical research closer to clinics and patients
- Take part in international organizations such as the Global Alliance for Genomics and Health (GA4GH), the European Genome Phenome Archive (EGA) and ELIXIR shaping the future of genomics across borders
- Widen your expertise with extensive possibilities of training programs, seminars and conferences
- Enjoy a flexible work environment with the opportunity to work from home part-time, balancing office presence and remote work
YOUR PROFILE
- Master's degree or equivalent qualification in bioinformatics, computational biology, biological science, computer science, physics, mathematics, engineering, or other fields, possibly with a PhD degree
- Demonstrated experience and expertise in the development of data management / data curation concepts is expected
- Expertise in NGS, (human) omics data, data security and data privacy is beneficial, as is communicating results and ideas to colleagues and (inter)national collaboration partners
- Proficiency with UNIX-based systems and relevant programming languages such as Python or R is a prerequisite
- Experience with the development and implementation of structured metadata using modeling languages such as LinkML and JSON Schema is beneficial
- Expertise in software development and cloud computing is beneficial
- Excellent collaboration and communication skills in English
The ideal applicant should have demonstrated the ability to work independently and creatively. The candidate should have excellent communications skills and be able to articulate clearly the technical needs, set clear goals and work within an interdisciplinary setting, communicating with other partners.
WE OFFER
Excellent framework conditions: state-of-the-art equipment and opportunities for international networking at the highest level
30 days of vacation per year
Flexible working hours
Remuneration according to TV-L incl. occupational pension plan and capital-forming payments
Possibility of mobile work and part-time work
Family-friendly working environment
Sustainable travel to work: subsidized Germany job ticket
Unleash your full potential: targeted offers for your personal development to further develop your talents
Our Corporate Health Management Program offers a holistic approach to your well-being
ARE YOU INTERESTED?
Then become part of the DKFZ and join us in contributing to a life without cancer!
Dr. Ivo Buchhalter
Phone: +49 (0)6221/42-3566
We are convinced that an innovative research and working environment thrives on the diversity of its employees. Therefore, we welcome applications from talented people, regardless of gender, cultural background, nationality, ethnicity, sexual identity, physical ability, religion and age. People with severe disabilities are given preference if they have the same aptitude.

- 2024-08-02
The newly founded "German Competence Centre Cloud Technologies for Data Management and Processing (de.KCD)", an association of the eight partners of the de.NBI Cloud, focuses primarily on teaching skills in the areas of workflows, cloud computing, data management and data processing in cloud computing environments. In close cooperation with de.NBI and ELIXIR, sustainable training materials are to be developed for researchers from various disciplines and teached in various courses or provided as online self-study units. For this task, we are looking for an experienced and motivated Research Associate with experience in (bio)informatics and with an interest in knowledge transfer in computer-aided (life)sciences.
Your tasks in detail:
- Design and definition of a sustainable process for the life cycle of training materials
- Design and development of high-quality training material embedded in a structured training programme
- Organisation and implementation of training courses and workshops and cooperation in this area with the project partners
- Support in the creation of information materials for the external presentation of the project
- Establishment of an internal monitoring and reporting system to record relevant key performance indicators for academic achievements
- Supporting project management in the organisation of project events and reporting
- Regular dialogue with internal and external project partners
- Active awareness of the latest developments in computational research, in particular using cloud computing technologies and Data Science
Your Profile:
- Scientific university degree (Master`s/Diploma) related to the life or natural sciences or related fields, preferably with a doctorate/Phd studies
- Practical experience in data-driven science (e.g. data analysis, applied statistics, visualisation, etc.)
- Strong interest, experience and/or formal qualifications (e.g. Carpentries, RStudio instructor certification, etc.) in teaching computer-based research skills in targeted training courses
- Working knowledge of programming/scripting languages such as Python, R, Bash and concepts of scientific data processing such as version control, cloud computing, cluster computing, workflow management, data literacy, etc.
- Very confident with Linux/Unix
- Knowledge of sequence analysis and metagenomics, especially in the processing of NGS data is an advantage
- Strong interpersonal skills and ability to work effectively in a team
- Strong communication skills, verbal and written expression in German and English
- Enjoy working in an interdisciplinary, science-orientated environment at the interface between science, teaching, administration, politics and business
- Willingness to travel and actively participate in conferences, networking events and project meetings
Our Offer:
We work on highly topical socially relevant issues and offer you the opportunity to play an active role in shaping change! We support you in your work through:
- Highly interesting, interdisciplinary, varied work in a highly topical subject area
- Participation in project meetings and conferences
- Strong support and mentoring for building a future career in academia and/or industry
- Further development of your personal strengths, e.g. through an extensive range of training courses; a structured further training programme and networking opportunities especially for PhD students via JuDocS, the Jülich centre for PhD students and supervisors:https://www.fz-juelich.de/judocs
- Optimal conditions for a good work-life balance and a family-conscious company policy
- The possibility to work flexibly (in terms of location), e.g. in a home office, is generally given after consultation and in accordance with the tasks and (on-site) appointments at hand
- Academic achievements and a company pension scheme
- Flexible working hours in a full-time position (39 hours/week) with the option of slightly reduced working hours
- 30 days holiday plus all bridging days and always off duty between Christmas and New Year
- Targeted services for international employees, e.g. through our International Advisory Service
We offer you an exciting and varied role in an international and interdisciplinary working environment. The position is initially for a fixed term of 2 years, with possible long-term prospects. Salary and social benefits will conform to the provisions of the Collective Agreement for the Public Service (TVöD-Bund), pay group EG 13, depending on the applicant’s qualifications and the precise nature of the tasks assigned to them.
In addition to exciting tasks and a collaborative working atmosphere at Jülich, we have a lot more to offer: https://go.fzj.de/benefits
Place of employment: Bielefeld
We welcome applications from people with diverse backgrounds, e.g. in terms of age, gender, disability, sexual orientation / identity, and social, ethnic and religious origin. A diverse and inclusive working environment with equal opportunities in which everyone can realize their potential is important to us.

- 2024-06-19
The new de.NBI Quarterly Newsletter, has been published. It reports on main de.NBI activities since March 2024.
Main topics:
- 3rd BioHackathon Germany – Call for project proposals open
- de.NBI/ELIXIR-DE All Hands Meeting - Save the date
- Re-activation of de.NBI working groups
- Reviews of de.NBI Spring School 2024, BrAPI Hackathon, and ELIXIR AHM 2024
- ...and much more!

- 2024-06-14
IT Center (ZDV), High Performance und Cloud Computing Group
Bewerbungsfrist: 14.07.2024
The High Performance und Cloud Computing Group at the IT Center (ZDV) of the Eberhard Karls Universität Tübingen seeks to fill one project position at the earliest possible date for
Scientific Staff (Wissenschaftliche Mitarbeiter (m/w/d; bis zu E 13 TV-L, 100%))
ZDV-09-24
The University of Tübingen is a central partner of the German Network for Bioinformatics Infrastructure (de.NBI) and provides a cloud infrastructure for computation, along with the Universities of Bielefeld, Freiburg, Gießen, Heidelberg, the DKFZ, Charité - BIH and the Research Centre Jülich (https://cloud.denbi.de/). The position focuses on cloud operations and development, collaboration with other cloud sites, and cloud user support.
As part of the de.NBI Cloud team you will be working on:
- Conception, procurement and operation of the hardware infrastructure
- Integration and operation of heterogenous storage resources
- Integration and operation of federated compute resources
- Integration and operation of network components
- Authentication and authorization
- Support of de.NBI Cloud users
- Handling of sensible research data
- Close collaboration with the project partners and the scientific community
- Active support for the broad user community for bioinformatical and technical questions
We expect from you:
- A scientific higher education degree (master, diploma or comparable) in bioinformatics, computer science or a related field
- Experience with an OpenStack-based cloud environment as user and/or administrator
The following competences are considered to be beneficial:
- Python expertise
- DevOps experiences with tools like Ansible, Terraform, Git, Kickstart
- Knowledge about network topologies and concepts like VLAN separation or Software-defined Networking
- Experience with software defined storage systems like Ceph or Quobyte
- Experience with Kubernetes or other container orchestration
- Experience with information security management systems, ideally including certification processes (ISO 27001)
- Experience with Monitoring tools like Prometheus, Grafana or Zabbix
- Team oriented but self-reliant working habits
- Professional interaction with heterogenous user groups
- High motivation, flexibility and team spirit
- Highly proficient in both spoken and written English
- Experience with multidisciplinary scientific working habits
We offer:
- A thorough initial training phase and continuous further training
- A stimulating working environment including space for personal initiative
- Work within a likeable and congenial team
- The opportunity to gain experience in all areas of cloud operation
- Flat hierarchies and the encouragement of own ideas
- Flexible working hours
- Job ticket
- University sport program
The position is initially limited until 31.12.2027. The university seeks to raise the number of women in research and teaching and therefore urges qualified women academics to apply for these positions. Equally qualified applicants with disabilities will be given preference. The employment will be carried out by the central administration of the University of Tübingen.
Have we caught your interest? Please send your application including cover letter, CV, resume and a copy of the certificate of your highest professional qualification, refereeing to reference number ZDV-09-24 until July 14th, 2024 to the director of the ZDV, Prof. Dr. Thomas Walter, 72074 Tübingen, Wächterstraße 76 or in electronic from to
Dr. Jens Krüger (

- 2024-05-22
Save the Date for the Fellowship of the Data Meeting 2025!
01 -03 April 2025
Face to face event in Jena with conference fee and: Keynotes! Discussions! Networking! Fun!
For everyone working as Data Steward*, feeling like one, or interested in the topic!
* Data Steward = RDM professional, trainer, data manager, curator, and all similar roles promoting research data management for scientists of any field and discipline from all nations.
3 Days, 3 Main Topics:
- Data Steward* Newbie Jumpstart: Overview of Landscape, Stakeholders, Support & Networks
- The Life of a Data Steward*: Tips & Tricks, Fails, Task Diversity
- Changing Research Data Management Culture: What, Why, How?
Further information will follow – like & subscribe to our newsletter mailing list: https://www.listserv.dfn.de/sympa/info/datafellows
Note that there will be no event in 2024. Please feel free to forward this Save the Date for 2025 to anyone interested! Please feel free to get in touch for questions or to support us Looking forward to see you all in person! Be a DataFellow!
Your Fellowship-Orga Team Cora (TKFDM), Nils (de.NBI/ELIXIR-DE), Elena (FAIRAgro), Jeanne (Leibniz FLI)
[Event formerly also titled RDM Trainer Network Meeting]

- 2024-05-13
Position: DevOps Engineer / Cloud Specialist
Department: German Human Genome-Phenome Archive (GHGA)
Code number: 2024-0150
The German Cancer Research Center is the largest biomedical research institution in Germany. With more than 3,000 employees, we operate an extensive scientific program in the field of cancer research.
The German Cancer Research Center is seeking for the recently launched infrastructure initiative German Human Genome-Phenome Archive (GHGA) a DevOps engineer / cloud specialist.
GHGA is part of the national program for research data infrastructures (NFDI). GHGA will act as a national node within the federated European Genome-Phenome Archive. This infrastructure will support genomics data hubs with software tools for secure data/metadata storage, interactive data portals with data visualization, and streamlined data deposition and acquisition solutions. The GHGA main office, which is coordinated by Prof. Oliver Stegle, is located at DKFZ in close coordination with the European Molecular Biology Laboratory (EMBL).
In order to develop the GHGA software platform into a state-of-the-art infrastructure for the secure exchange of genome data, we are looking for a DevOps engineer / cloud specialist to join the team as soon as possible.
Job description:
We are looking for a team member with substantial experience and/or interest in developing, deploying, and maintaining the cloud platforms for GHGA infrastructure. The successful candidate will be part of an interdisciplinary research, development, and data management team to develop and apply a diverse range of state-of-the-art technologies.
Your responsibilities:
- Build a state-of-the-art infrastructure for deployment of services across multiple data centers
- Design and implement scalable solutions based on modern cloud frameworks and technologies such as Kubernetes, Service & Event Meshes
- Build hybrid cloud solutions that span multiple private or public cloud providers and include edge computing scenarios
- Implementing system security measures and performing penetration testing
- Supporting developers in an agile setting to productionalize their software and make it ready for continuous deployment
- Supporting system administrators of participating academic institutions in migrating from classical HPC environments (Slurm, LSF) to cloud-based settings
Your possibilities:
- Expand your technical skills through curiosity-driven projects in an academic setting that values innovation and supports exploration of new ideas and technologies
- Get exposed to a plethora of fields ranging from AI-guided clinical decision making to modern web development
- Work with an interdisciplinary team of experts bringing state-of-the-art biomedical research closer to clinics and patients
- Take part in international organizations such as the Global Alliance for Genomics and Health (GA4GH), the European Genome-Phenome Archive (EGA) and ELIXIR shaping future of genomics across borders
- Widen your expertise with extensive possibilities of training programs, seminars and conferences
Requirements:
- Bachelor's or master’s degree in computer science or similar technical background
- Extensive experience in building and deploying Docker containers
- Familiarity with infrastructure as code frameworks such as Ansible or TerraForm
- Experience in describing container relations as code using e.g. Helm Charts or Docker Compose
- Experience with CI/CD pipelines such as GitHub Actions
- Experience and/or strong interest in working with Kubernetes clusters
- First hand experience with private OpenStack-based clouds, public providers such as AWS, Azure, and GCS, familiarity with GitOps services like Argo CD, Flux CD, or Jenkins X is a plus
- Enjoy working in an interdisciplinary team and talking to software developers, system administrators, as well as domain experts
- Enjoy working with modern technologies and staying up to date with new tech trends
The ideal applicant should have demonstrated the ability to work independently and creatively. The candidate should have excellent communications skills in English and be able to articulate clearly the technical needs, set clear goals and work within an interdisciplinary setting, communicating with other partners.
We offer:
- Excellent framework conditions: state-of-the-art equipment and opportunities for international networking at the highest level
- Remuneration according to TV-L incl. occupational pension plan and capital-forming payments
- 30 days of vacation per year
- Flexible working hours
- Possibility of mobile work and part-time work
- Family-friendly working environment, e.g. parent-child room, advisory services caring for elderly relatives
- Sustainable travel to work: subsidized Germany job ticket
- Unleash your full potential: targeted offers for your personal development to further develop your talents
- Our Corporate Health Management Program offers a holistic approach to your well-being
Important notice:
The DKFZ is subject to the regulations of the Infection Protection Act (IfSG). As a consequence, only persons who present proof of immunity against measles may work at the DKFZ.
Earliest Possible Start Date: as soon as possible
Duration: The position is initially limited to 2 years with the possibility of prolongation.
Application Deadline: 30.05.2024
Contact:
Dr. Jan Eufinger
Phone +49 (0)6221/42-3466
Please note that we do not accept applications submitted via email.

- 2024-05-13
The Department of Computer Science is opening a position for a Systems Engineer / DevOps Engineer (m/f/d), bridging development and operations functions within one of the most popular and well-established data analysis platforms for life science. The engineer will be responsible for implementing, supporting, and documenting several large-scale application systems. This role includes but is not limited to, the installation, modification, testing, and upgrades of applications, operating systems, file systems, hardware, and cloud environments. The work will be carried out together with other administrators.
Who we are
The European Galaxy team is a core group within the worldwide Galaxy project. We are an open, passionate, and supportive research group at the University of Freiburg highly involved in many software projects related to the global Galaxy community, including leading the Bioconda and BioContainers projects and developing state-of-the-art computational workflows for genomics, metagenomics, transcriptomics, and pharmacogenomics data.
Based in Freiburg, usegalaxy.eu is the science gateway, providing an open-access computational infrastructure for accessing and running a vast catalog of data resources, tools, and workflows. It is used by nearly 100,000 users worldwide.
Your opportunities
This position will allow the postholder to interact with an interdisciplinary bioinformatics, data science, and software development team, working with cutting-edge cloud infrastructure across Europe and engaged in advanced topics such as distributed heterogeneous computing, encrypted data processing, or smart scheduling of jobs across federated resources.
Your Tasks
- Development & maintenance of the Galaxy server and related infrastructure, e.g., TIaaS (Training Infrastructure as a Service), Pulsar-Network, and the Beacon Service
- Work within the team of Admins to design and improve developed software and deployment pipelines
- Conceptualization of update procedures and software integrations in the running system
- Implementing and maintaining best security practices and adhering to our information security policies and industry standards
- Staying current with industry trends and emerging technologies, e.g., virtualization and infrastructure technologies, automatization, and CI/CD pipelines
- Proactively identifying opportunities to enhance the Galaxy Europe instance as well as general deployment guides for other Galaxy instances
- Integrating infrastructural components like various storage systems and special-purpose computation nodes containing accelerators like GPUs
- Community support and training
- Development of documentation and training material for Galaxy admins and automation workflows
- support of services administrators
- give workshops and train other administrators in live sessions
Your Profile
We are looking for a dedicated and motivated team player with strong communication skills and a good command of the English language.
The successful applicant will be an expert Linux system administrator with proficient knowledge of Python, Git, and SQL databases and a Bachelor or Master in Informatics or equal qualification. He/she is an analytical thinker, problem-solver and reliable, with a strong desire to learn and collaborate with the team.
Additional skills that are advantageous:
- several years experience
- Experience building containerized applications
- Experience configuring continuous integration and continuous delivery (CI/CD) systems
- Proficiency using configuration management tools (Terraform, Ansible)
- Familiarity with cloud technologies (e.g. OpenStack)
- Experience in high-performance computing environments
What we offer
Located in a region that connects Germany, France, and Switzerland, our institute offers an international research environment with outstanding infrastructure facilities and a positive working atmosphere that places a high value on work-life balance.
- A team of admins to work with
- Professional training and development opportunities
- Challenging tasks in European-wide distributed projects in a modern working environment
- Flexible working hours and trust-based working time
- 30 days holiday a year
- Reduced-cost public transport within the local area
- Family-friendly offers (affiliated daycare center at the institute, family service)
This position is open immediately and funded for two years, with the possibility of extension. The salary will be determined in accordance with TV-L up to E13 if the personal and collective bargaining requirements are met.
More information about the European Galaxy project is available at https://galaxyproject.eu.
Ready to join our team?
Please submit your complete application documents, including a 1-page motivation summary, CV and contact details.
We are particularly pleased to receive applications from women for the position advertised here.
Application
Please send your application in English including supporting documents mentioned above citing the reference number 00003737, by 10.06.2024 at the latest. Please send your application to the following address in written or electronic form:
University of Freiburg
Institut für Informatik, Bioinformatik
Dr. Björn Grüning
Head of Freiburg Galaxy Team
Georges-Köhler-Allee 106
79110 Freiburg
Germany
E-Mail:
For further information, please contact Frau Dr. Anika Erxleben-Eggenhofer on the phone number +49 761 203-54130 or E-Mail

- 2024-04-08
IT Center (ZDV), High Performance und Cloud Computing Group
Bewerbungsfrist: 14.04.2024
The High Performance und Cloud Computing Group at the IT Center (ZDV) of the Eberhard Karls Universität Tübingen seeks to fill one project position at the earliest possible date for
Scientific Staff (Wissenschaftliche Mitarbeiter (m/w/d; bis zu E 13 TV-L, 100%))
ZDV-07-24
The University of Tübingen is a central partner of the German Network for Bioinformatics Infrastructure (de.NBI) and provides a cloud infrastructure for computation, along with the Universities of Bielefeld, Freiburg, Gießen, Heidelberg, the DKFZ, Charité - BIH and the Research Centre Jülich (https://cloud.denbi.de/). The position focuses on cloud operations and development, collaboration with other cloud sites, and cloud user support.
As part of the de.NBI Cloud team you will be working on:
- Conception, procurement and operation of the hardware infrastructure
- Integration and operation of heterogenous storage resources
- Integration and operation of federated compute resources
- Integration and operation of network components
- Authentication and authorization
- Support of de.NBI Cloud users
- Handling of sensible research data
- Close collaboration with the project partners and the scientific community
- Active support for the broad user community for bioinformatical and technical questions
We expect from you:
- A scientific higher education degree (master, diploma or comparable) in bioinformatics, computer science or a related field
- Experience with an OpenStack-based cloud environment as user and/or administrator
The following competences are considered to be beneficial:
- Python expertise
- DevOps experiences with tools like Ansible, Terraform, Git, Kickstart
- Knowledge about network topologies and concepts like VLAN separation or Software-defined Networking
- Experience with software defined storage systems like Ceph or Quobyte
- Experience with Kubernetes or other container orchestration
- Experience with information security management systems, ideally including certification processes (ISO 27001)
- Experience with Monitoring tools like Prometheus, Grafana or Zabbix
- Team oriented but self-reliant working habits
- Professional interaction with heterogenous user groups
- High motivation, flexibility and team spirit
- Highly proficient in both spoken and written English
- Experience with multidisciplinary scientific working habits
We offer:
- A thorough initial training phase and continuous further training
- A stimulating working environment including space for personal initiative
- Work within a likeable and congenial team
- The opportunity to gain experience in all areas of cloud operation
- Flat hierarchies and the encouragement of own ideas
- Flexible working hours
- Job ticket
- University sport program
The position is initially limited until 31.12.2027. The university seeks to raise the number of women in research and teaching and therefore urges qualified women academics to apply for these positions. Equally qualified applicants with disabilities will be given preference. The employment will be carried out by the central administration of the University of Tübingen.
Have we caught your interest? Please send your application including cover letter, CV, resume and a copy of the certificate of your highest professional qualification, refereeing to reference number ZDV-07-24 until April 14th, 2024 to the director of the ZDV, Prof. Dr. Thomas Walter, 72074 Tübingen, Wächterstraße 76 or in electronic from to
Dr. Jens Krüger (

- 2024-03-26
The new de.NBI Quarterly Newsletter, has been published. It reports on main de.NBI activities in Q1 2024.
Main topics:
- New de.NBI/ELIXIR-DE consortium – national contract signed
- de.NBI and ELIXIR-DE now led by Oliver Kohlbacher and Alexander Sczyrba
- FAIR training handbook published by the ELIXIR FAIR training focus group
- ...and much more!

- 2024-03-20
To encourage the re-use of your training material it helps to ensure Findability, Accessibility, Interoperability and Reusability (FAIR). However, it is often not straightforward to apply the FAIR principles to training material. de.NBI and other ELIXIR members just released a handbook to guide you in this process. This comprehensive book provides examples and guidelines to support anyone that is involved in the creation of training materials, such as trainers, training coordinators and Train-the-Trainer instructors. The book consists of ten chapters that treat amongst others how to choose the material formats, annotate materials with metadata, get a persistent identifier and boost discoverability. It can serve as a reference to learn about general best-practices or to quickly look up specific how-tos. This book is an ongoing effort; databases, registries and best practices are constantly evolving. Therefore, any feedback or contributions from the community are warmly welcomed.
Check it out! https://elixir-europe-training.github.io/ELIXIR-TrP-FAIR-training-handbook/


- 2024-01-17
Opportunity: We are looking for candidates to fill an open staff scientist position with a strong background in data science, data engineering or software development to facilitate and assist with the development of ProteomicsDB. ProteomicsDB is an internationally well-reputed publicly available database that provides information about proteins and other bio-molecules initially centered around human biology. It is designed to integrate data from mass spectrometry-based experiments and other sources to offer a comprehensive view of proteomes. We are continuously expanding the types of data stored in as well as the number of organisms covered. Researchers and scientists can use ProteomicsDB to explore and analyze protein-related data for a better understanding of cellular processes, functions, and disease mechanisms. ProteomicsDB is built on the in-memory system SAP HANA using IBM Power 8/9 hardware.
The successful candidate will become a key member of a strong interdisciplinary team of bioinformatician that focus on the development and application of novel approaches in proteomic research. Depending on the applicant’s skills, this position will strengthen our ongoing development of ProteomicsDB focusing on the backend or frontend development. In light of this, the most important aspects of the job description are:
- Service engineering and infrastructure maintenance for ProteomicsDB
- Full-stack development to enhance ProteomicsDB
- Data import, maintenance, and integration of proteomics, lipidomics, or metabolomics data
Requirements: Candidates must hold a master’s degree in Data Engineering, Data Science, Bioinformatics, Informatics, or a related discipline. Essential skills include a sound understanding of Linux and related systems, software development standards, and hardware management. Additional beneficial skills include a theoretical knowledge of and practical skills in statistical analysis, data integration, backend or frontend programming and database design. Interest in understanding technologies e.g. proteomics and metabolomics using mass spectrometers is expected. We are looking for a self-motivated and broadly interested individual with high potential and a strong sense of responsibility. Flexibility and the ability to work in a fast-paced environment on multiple scientific and infrastructure projects are essential. Good inter-cultural and inter-personal communication skills as well as the ability to present in English are also important.
Our offer: You will join a young and highly motivated team of interdisciplinary bioinformaticians that use the latest proteomic approaches to gain insight into the biological processes that govern life. You will further be connected to the Munich Data Science Institute (MDSI) at the Technical University of Munich (TUM) to foster and facilitate exchange. You will also be connected to the SAP University Competence Center (UCC) at the TUM to assist in the hardware/software maintenance of ProteomicsDB, with the option to receive a proof of work with SAP HANA. TUM is one of the best academic institutions in Germany and offers a stimulating work environment and excellent future perspectives.
The position is limited until December 2025 and available as soon as possible. Remuneration is in accordance with the TV-L E13 according to the professional qualification.
Equal opportunity: TUM is aiming to increase the proportion of women and, therefore, expressly welcomes applications from women. The position is also suitable for severely disabled persons. Severely disabled applicants will be given preference if their suitability, qualifications and professional performance are otherwise essentially equal.
Application process: Applicants should send a dossier containing a motivational statement (max. one page), a curriculum vitae summarizing qualifications and experience, copies of degrees and transcripts of study records, names and the email addresses of at least one referee as a single PDF document to Mathias Wilhelm (via mail
Data Protection Information: When you apply for a position with the Technical University of Munich (TUM), you are submitting personal information. With regard to personal information, please take note of the Datenschutzhinweise gemäß Art. 13 Datenschutz-Grundverordnung (DSGVO) zur Erhebung und Verarbeitung von personenbezogenen Daten im Rahmen Ihrer Bewerbung. (data protection information on collecting and processing personal data contained in your application in accordance with Art. 13 of the General Data Protection Regulation (GDPR). By submitting your application, you confirm that you have acknowledged the above data protection information of TUM.

- 2023-11-22

| Issue 04/2023 | ||||||||||||||||||||||||||||||
Next Event: 23 Nov 2023, Annual Meeting of the de.NBI Industrial Forum |
||||||||||||||||||||||||||||||
 The 4th Annual Meeting will cover industrial and academic stakeholder presentations on the theme Data Handling in the Life Sciences. This event will be held virtually on 23 November 2023 from 1-5 pm (CET). The 4th Annual Meeting will cover industrial and academic stakeholder presentations on the theme Data Handling in the Life Sciences. This event will be held virtually on 23 November 2023 from 1-5 pm (CET).Presentations about de.NBI Cloud, an introduction to GAIA-X based Health-X dataLOFT, and data handling in the NFDI4Microbiota consortium will kick off the meeting. Industrial use cases will feature:
Further information, agenda, abstracts, and registration here.
We look forward to welcoming you to the 4th Annual Meeting of the de.NBI Industrial Forum. |
||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
| Andrew Smith as interim ELIXIR Director
Niklas Blomberg will step down as ELIXIR's Director at the end of this year to join the Innovative Health Initiative (IHI) as Executive Director. The ELIXIR Board has approved the recommended candidate for the Director position, and details about the new Director will be confirmed soon. In the meantime, the Board appointed Andrew Smith as interim Director, for an initial term of six months, from December 2023 to May 2024. Alexander Goesmann elected Chair of the ELIXIR Board The ELIXIR Staff Exchange Programme aims to facilitate meaningful networking and engage with experts from various ELIXIR nodes, fostering cross-disciplinary collaboration.ELIXIR's commitment to staff exchanges reflects its recognition of the value in nurturing connections within the scientific community. By facilitating these opportunities, ELIXIR empowers researchers to collaborate on a global scale, contributing to the growth and progress of the bioinformatics field. In 2023, members from ELIXIR Germany particitpate in four projects:
ELIXIR launched the ELIXIR Impact Toolkit |
||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
|
Since the initial publication of Galaxy: a platform for interactive large-scale genome analysis in September 2005, a multitude of subsequent papers, authored by both the Galaxy team and its users, have contributed to Galaxy's extensive citation record, exceeding 12,000 globally. This pivotal paper marked the establishment of the global Galaxy community, significantly influencing computational biology and related fields. Congratulations to the inventors, contributors, and supporters on 18 years of success! Here's to looking forward to another 18 years of continuous innovation and collaboration! More details see here. |
||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
|
Science communication of a different kind |
||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
|
Recent updates of services:
|
||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
| The next de.NBI training events are announced on www.denbi.de/training.
Upcoming scheduled events:
|
||||||||||||||||||||||||||||||
Impressum |
||||||||||||||||||||||||||||||
|
Responsible for contents:
T. Dammann-Kalinowski, I. Maus, D. Wibberg
de.NBI - German Network for Bioinformatics Infrastructure, Forschungszentrum Juelich GmbH - IBG-5, Branch Office at Bielefeld University, 33594 Bielefeld, Germany Phone: +49-(0)521-106-8758
Fax: +49-(0)521-106-89046 Email: Web:https://www.denbi.de
Twitter: #denbi, @denbiOffice LinkedIn: https://www.linkedin.com/company/de-nbi For questions and remarks please contact Subscription
If you are interested in receiving regular updates on de.NBI - German Network of Bioinformatics Infrastructure, please subscribe to the de.NBI Quarterly Newsletter here. The mailing list will exclusively be used to distribute the de.NBI Quarterly Newsletter.
To unsubscribe from the de.NBI Quarterly Newsletter, please use the unsubscribe link. The de.NBI Quarterly Newsletter is a service of de.NBI - German Network for Bioinformatics Infrastructure for members, partners and interested public. All photos are copyright of the de.NBI administration office unless marked otherwise. |











































 de.NBI will continue its very successful school training program with the de.NBI Spring School 2024 on Bioinformatics for Microbial Omics to be held in Bielefeld from 18-22 March, 2024. It will be organized by the CAU and the Bielefeld-Gießen Resource Center for Microbial Bioinformatics (BiGi).
de.NBI will continue its very successful school training program with the de.NBI Spring School 2024 on Bioinformatics for Microbial Omics to be held in Bielefeld from 18-22 March, 2024. It will be organized by the CAU and the Bielefeld-Gießen Resource Center for Microbial Bioinformatics (BiGi). Prof. Alexander Goesmann (Germany) from the Justus-Liebig University Giessen was elected the Chair of the
Prof. Alexander Goesmann (Germany) from the Justus-Liebig University Giessen was elected the Chair of the 
 The
The 

 Alexander Sczyrba appointed Director of section IBG-5 "Computational Metagenomics" at FZJ
Alexander Sczyrba appointed Director of section IBG-5 "Computational Metagenomics" at FZJ Johanna Nelkner, Service Coordinator at the de.NBI administration office, has won this year's
Johanna Nelkner, Service Coordinator at the de.NBI administration office, has won this year's 

