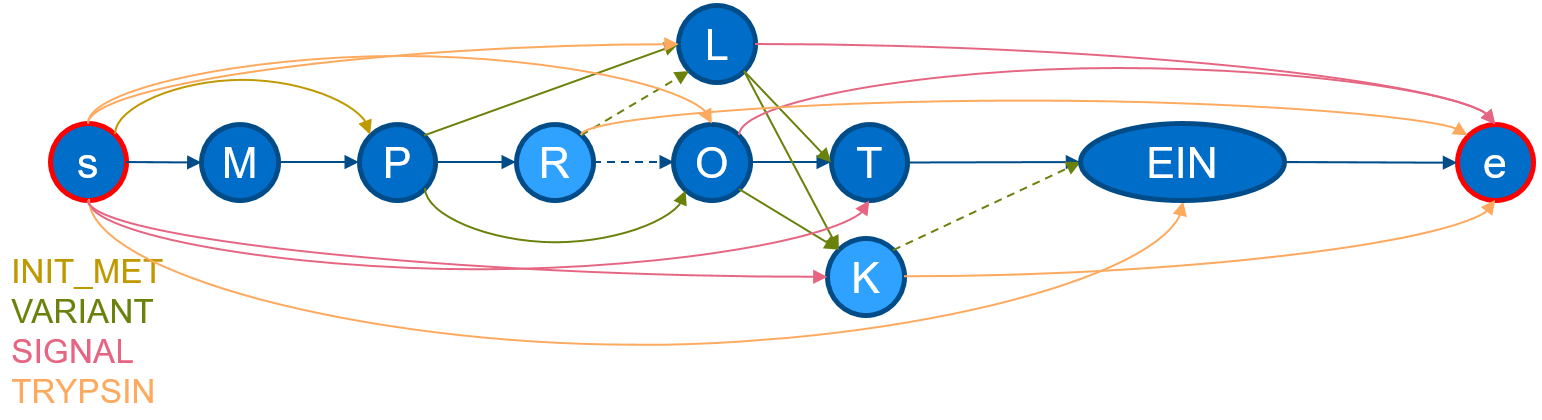
ProtGraph
ProtGraph is a Python package that converts protein entries from UniProtKB into protein graphs (directed acyclic graphs). It parses UniProtKB SP-EMBL-style entries (e.g. .txt / .dat) including the canonical sequence and rich feature annotations such as isoform sequences, specifically cleaved peptides (e.g. signal peptides, propeptides), and variational changes (e.g. variants, mutations, sequence conflicts). Depending on the configuration, ProtGraph generates graphs that represent all (or selected) combinations of these features, and can be extended with digestion information and post-translational modifications. This enables estimation of theoretical protein/peptide search spaces for species (e.g. human, mouse) or even the complete UniProtKB.
Key benefits
- Graph-based representation of protein sequence variability: models isoforms, cleavage products, variants, and other UniProtKB features in a unified directed acyclic graph.
- Configurable feature inclusion: generate graphs that include all annotations or focus on selected feature types, depending on the analysis goal.
- Supports proteomics search-space estimation: helps assess theoretical protein/peptide search spaces across organisms and datasets.
- Extensible with digestion and PTMs: optionally enriches graphs to better reflect downstream proteomics workflows.
- Interoperable outputs: exports graphs and sequences in multiple formats, enabling inspection with external graph tools and reuse in downstream pipelines.
Applications
- Generation of protein graphs capturing sequence variants, isoforms, and proteolytic processing
- Estimation of theoretical protein/peptide search spaces for specific species or UniProtKB-wide analyses
- Export of custom-tailored FASTA files for proteomics database search workflows
- Graph export and complexity inspection using external visualization/graph analysis tools
- End-to-end workflow usage demonstrated via ProGFASTAGen
Intended use
ProtGraph is intended for researchers in proteomics and computational biology who want to represent protein sequence diversity in a structured, configurable way and derive sequence resources for downstream analyses. Basic bioinformatics skills and command-line experience are recommended, since ProtGraph is a command line tool (CLI).
More information: ProtGraph on GitHub · ProtGraph on PyPI · ProtGraph on Bioconda