Eisenacher Group, Ruhr University Bochum, Service center: Bioinformatics for Proteomics – BioInfra.Prot
The overarching goal of “DeProVIDEO – Deep Learning for Protein VarIants DEtectiOn” is the detection of variants in the amino acid sequence of proteins in mass spectrometry data. The complete concept can be split into two parts, which both base on deep learning approaches.
Genetic variants can be the cause of specific diseases or give a predisposition to these, while often proteins are the actually afflicted biomolecules of a genetic variation. Changes in the genome can cause simple amino acid exchanges in corresponding proteins, but could also lead to more complex changes like insertions or deletions of longer sequences. While the genetic causes for these protein variants are known, tools to detect variants without a known genetic background will be developed in DeProVIDEO.
As the insertion of variants into a given protein database drastically increases the search space, problems occur during a statistical analysis especially considering the estimation of false positive rates of identifications. Therefore, the first part of the project addresses the development of a spectrum centric, instead of a global, false positive estimation of identified peptides. This will be approached by the application of spectra predictions for database annotated peptide sequences and variants using a deep neural network.
The second part of the project uses specific deep learning algorithms to identify peptide sequences of measured spectra without database information by so called de novo strategies. With this approach yet unknown variants could be identified, which might originate neither from genetic variants nor could be detected by other proteomic methods.
All created tools and models will be made publicly available to the proteomics community. For further information visit the website of the Medical Proteome Center.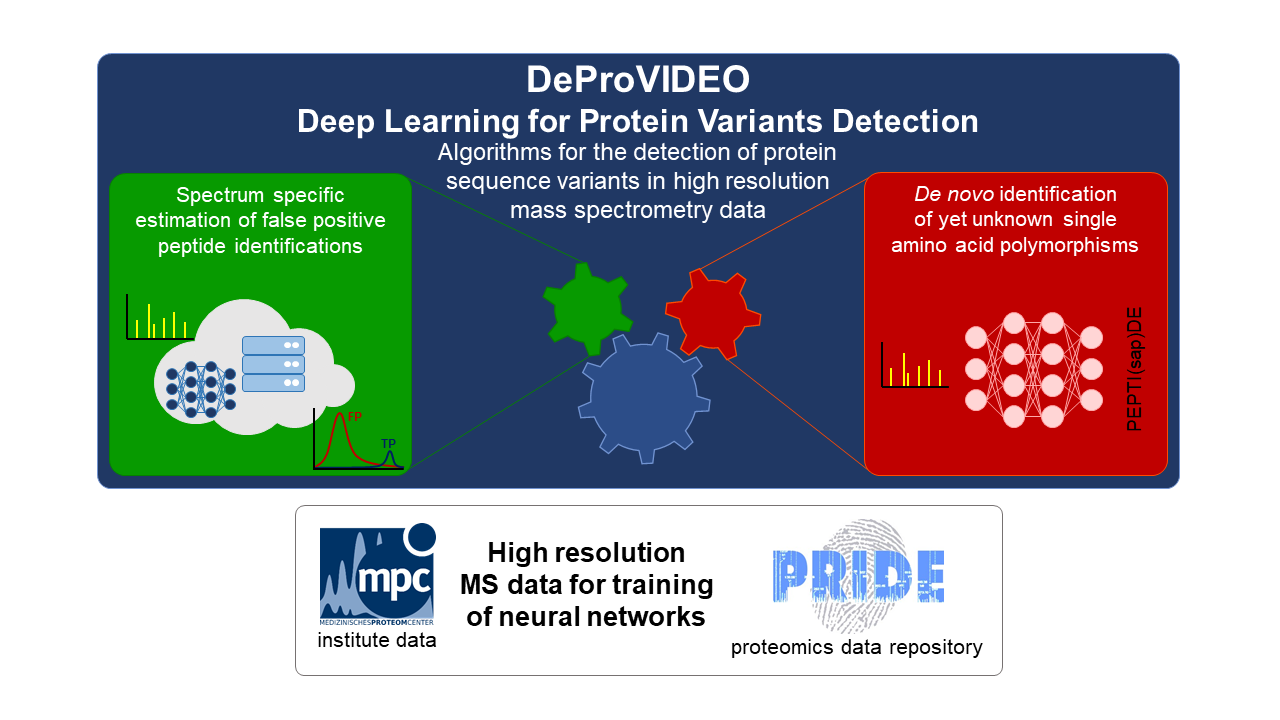
Search projects by keywords:

