
Genomics
-
Cardiovascular risk modelling
Eils Group, Hub for Innovations in Digital Health, Service center: Heidelberg Center for Human Bioinformatics – HD-HuB
In this project, we work towards an improved prevention and treatment of patients with established coronary heart disease (CHD). CHD is a major cause of morbidity and mortality. The risk landscape in patients with coronary disease is highly heterogenous, depending on their genetic background, clinical characteristics, cardiovascular risk factors and atherosclerotic disease status. With the availability of effective, but costly novel treatment options (such as PCSK9-inhibitors), there is great need for advanced cardiovascular risk prediction tools to stratify patients, guide the use of novel treatments and improve clinical trial design by selecting high-risk individuals. Here, we aim to improve and personalize prevention and treatment of coronary heart disease by developing data-driven, neural-network based tools for risk modelling and multi-modal data integration. We investigate representation learning techniques to identify latent factors across data modalities associated with risk and examine lifetime risk more closely with the aim of making individualized risk prediction using counterfactuals. The clinical environment at Charité in synergy with data-collection and patient management platforms allows for a prospective validation and clinical integration of our technologies.
For further information, visit ails lab.
 Search projects by keywords:
Search projects by keywords: -
Continuous Integration of RNAz, a bioinformatical research software for de novo detection of stable, conserved and functional noncoding RNAs in comparative genomics data
Stadler Group, Leipzig University, Service Center: RNA Bioinformatics Center - RBC
RNAz is a well established software to identify conserved RNA structures in genome-wide multiple sequence alignments. It has been widely used for more than a decade. RNAz makes use of the ViennaRNA package to compute secondary structures and their energies. Progress in the understanding of RNA folding, sequencing technologies, and alignment algorithms makes it necessary to restructure the RNAz software. In order to adapt RNAz to the expanded variations of energy models and to achieve longterm maintainability of this software we will devise automatized pipelines for the re-training of regression and decision models that are an essential part of RNAz and provide a repository of such models and corresponding datasets for the community.
For more information see RNA Bioinformatics Center (RBC)
Funded by: Federal Ministry of Education and Research 031A538B
Search projects by keywords:
-
Identification of new antimicrobial resistance targets by high-throughput deep learning
Goesmann Group, Gießen University, Service Center: Bielefeld-Giessen Resource Center for Microbial Bioinformatics - BiGi
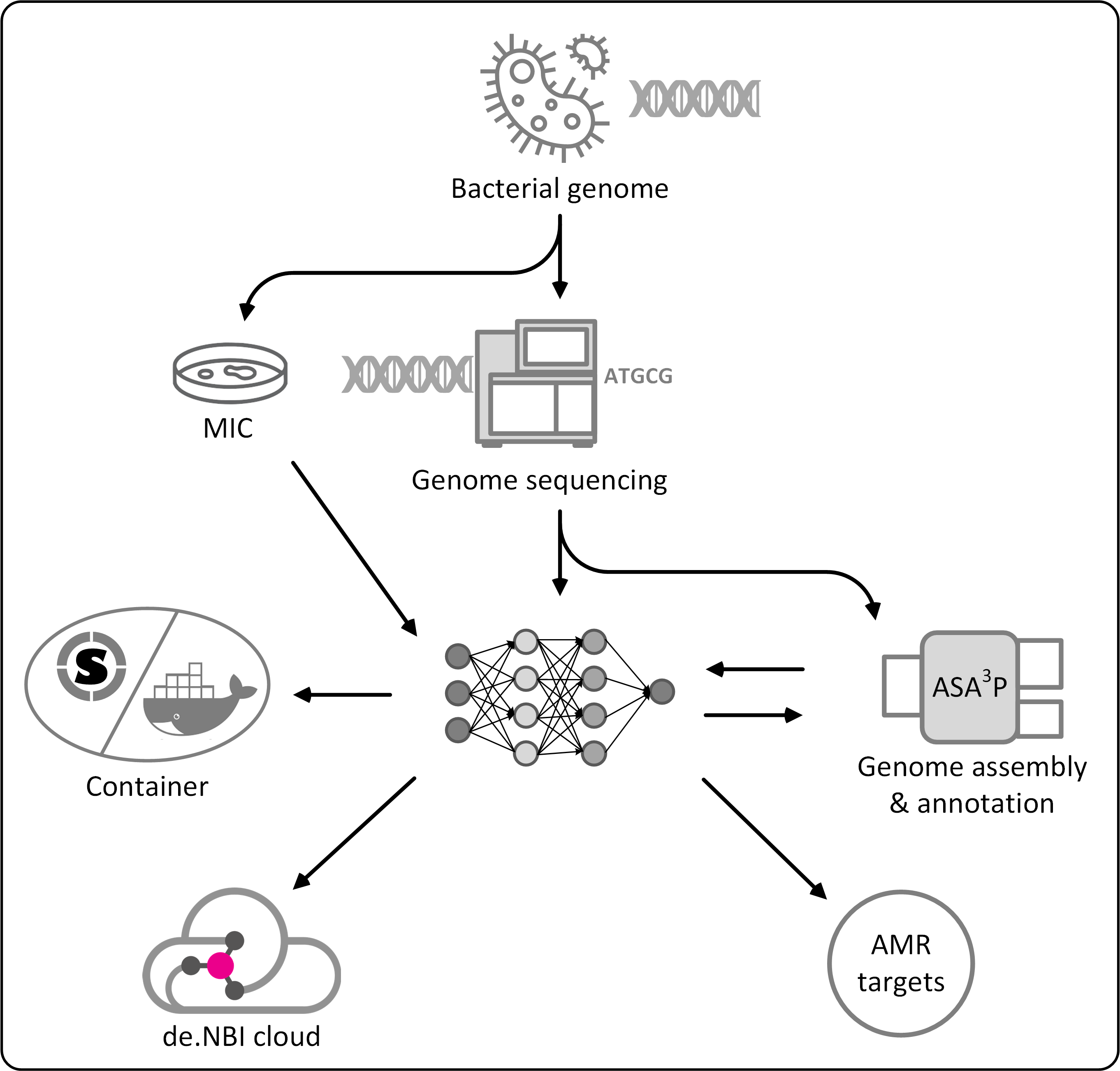
Antibiotic resistant bacteria have become a global severe threat for public health. Their surveillance and containment as well as the timely identification of new genetic determinants of antibiotic resistances are important tools to fight back emergent as well as established pathogens. Addressing these challenges, researchers from universities in Giessen and Marburg have started a new project called Deep-iAMR to combine expertise in medical microbiology, machine learning and cloud computing techniques. The aim of this new collaboration is to screen large amounts of genomic as well as phenotypic data for complex patterns between genomic and phenotypic data to find the genetic determinants of hard to detect or entirely new antimicrobial resistances. However, the application of modern artificial intelligence methodologies to find these patterns requires massive computing resources as well as specialized hardware, such as GPUs, tensors and FPGUs.
Researchers within the Deep-iAMR project take advantage of the cloud computing infrastructure of the de.NBI network which provides required computing resources as well as specialized hardware on demand. Resulting new methodologies and scalable tools will be shared with the scientific community and discovered new antibiotic targets will help to design necessary and urgently required new drugs to cure infections with highly-resistant pathogens.
For further information, please visit Deep-iAMR. To access the de.NBI Cloud visit de.NBI Cloud.
 Search projects by keywords:
Search projects by keywords: -
LAMarCK - LongitudinAI Multiomics Characterization of disease using prior Knowledge
Zeller Group, Saez-Rodriguez Group, Schlesner Group, Bork Group, and Korbel Group, Service center: Heidelberg Center for Human Bioinformatics – HD-HuB
The need for proper integration of molecular omics data from human individuals is arguably one of the biggest challenges in current biomedical research, as these data cover different functional aspects that facilitate basic research, but also reveal differences between human physiological states with medical relevance (e.g. disease versus healthy, different disease subtypes etc.). To tackle this challenge, we formed a team of researchers with complementary areas of expertise from different Heidelberg institutions, all associated with the de.NBI node HD-HuB, which delivers services on human genomics and human microbiomics. Both these fields rely on similar kinds of data, which are increasingly coming from longitudinal studies assessing multiple omics approaches. Thus, they face similar technical and methodological challenges - and closer communication, as well as integrative approaches bridging both fields hold great promise for improved diagnostic, prognostic and therapeutic approaches, as is already becoming apparent for type-2-diabetes onset.
The LAMarCK consortium on the one hand aims to devise generic approaches to integrate heterogeneous longitudinal multi-omics data across microbiome and host, on the other hand it will also develop frameworks that make use of domain-specific prior knowledge on the underlying biological mechanisms for improved interpretability of the results. Funded by the BMBF (grant no. 031L0181A), the consortium is applying and benchmarking state-of-the-art machine learning approaches for longitudinal multi-omics data from microbial and host cells and augmenting these by integration with biomolecular knowledge bases. These new approaches will help dissect the molecular mechanisms underlying human diseases, such as colorectal cancer.
Search projects by Keywords:
-
Omni-genetic phenotype models
Eils Group, Hub for Innovations in Digital Health, Service center: Heidelberg Center for Human Bioinformatics – HD-HuB
Genetic contributions to many health-related phenotypes like blood pressure and body mass index often arise from complex interactions between multiple genes. Thus, an understanding of how variants of different genes interact, is of great interest. We plan to make use of recent advances in the field of explainable machine learning to analyze data from large cohort studies. Starting with variants of individual genes, we want to model how they affect biological systems, for example DNA repair. Based on the combination of smaller specific subsystems, we want to predict the activity of more complex systems and repeat this process until the information of all genes is combined in one system describing the effect of all variants. At that point, the information on thousands of genes will be available in a very compressed representation, which will be the basis to predict phenotypes like blood pressure or other medically relevant phenotypes.
Additionally, this approach can be used to investigate which cellular systems in the model are affected by a given set of variants resulting in a specific phenotype. For example, for an individual with increased blood pressure and variants in multiple genes, this could be used to predict, whether the DNA repair system is involved in the increased blood pressure. Information like this can be used to check if the model makes its predictions in an intuitive way and can also be the starting point of new research, if strong connections are found.
For further information, please visit ails lab.

-
Scalable Curation and Genome Function Prediction by the aid of Artificial Intelligence
Overmann Group, Leibniz Institute DSMZ, Service center: Center for Biological Data – BioData
The rapidly increasing amount of data produced in science comes with new opportunities in analysis but also with new challenges in processing, interpreting, and storing data. The risk is high that large amounts of the data are only superficially analyzed and are being dumped on local storage without sufficient metadata enabling reuse of the data. Moreover, publication numbers rise quickly, but the data aggregated are difficult to access and not available for large scale analysis.
In a new approach (DiASPora project), the Leibniz Institute DSMZ is synthesizing information for bacterial species from diverse sources, together with the TIB Hanover, and ZB MED Cologne and publishes these in the database BacDive.
So far, the curation of scientific data is still a largely manual process that does not scale with the increasing amounts of data published every day. Therefore, workflows are developed that apply text mining combined with machine learning approaches including deep neural networks to automatically extract data for more than 150 microbial data types from literature. To attain high quality and correctness of the extracted data, a manual curation feedback loop will be integrated into the text mining pipeline, which enables to train the AI and thereby successively improve the quality.
The increasing availability of genomic data enables the prediction of bacterial traits based on genome annotation data. To achieve high quality in function prediction, classifiers are trained and tested with phenotypic data from BacDive and standardized genome annotations. An iterative optimization process including manual curation will retrieve the best models for a number of traits, allowing us to make predictions with a high confidence level for so far poorly studied bacteria.
All data that reach a high level of confidence will be standardized and published in BacDive (de.NBI tool) in a machine-interpretable format, ready for reuse and large-scale analysis.
For further information visit the following websites for DiASPora project and BacDive.
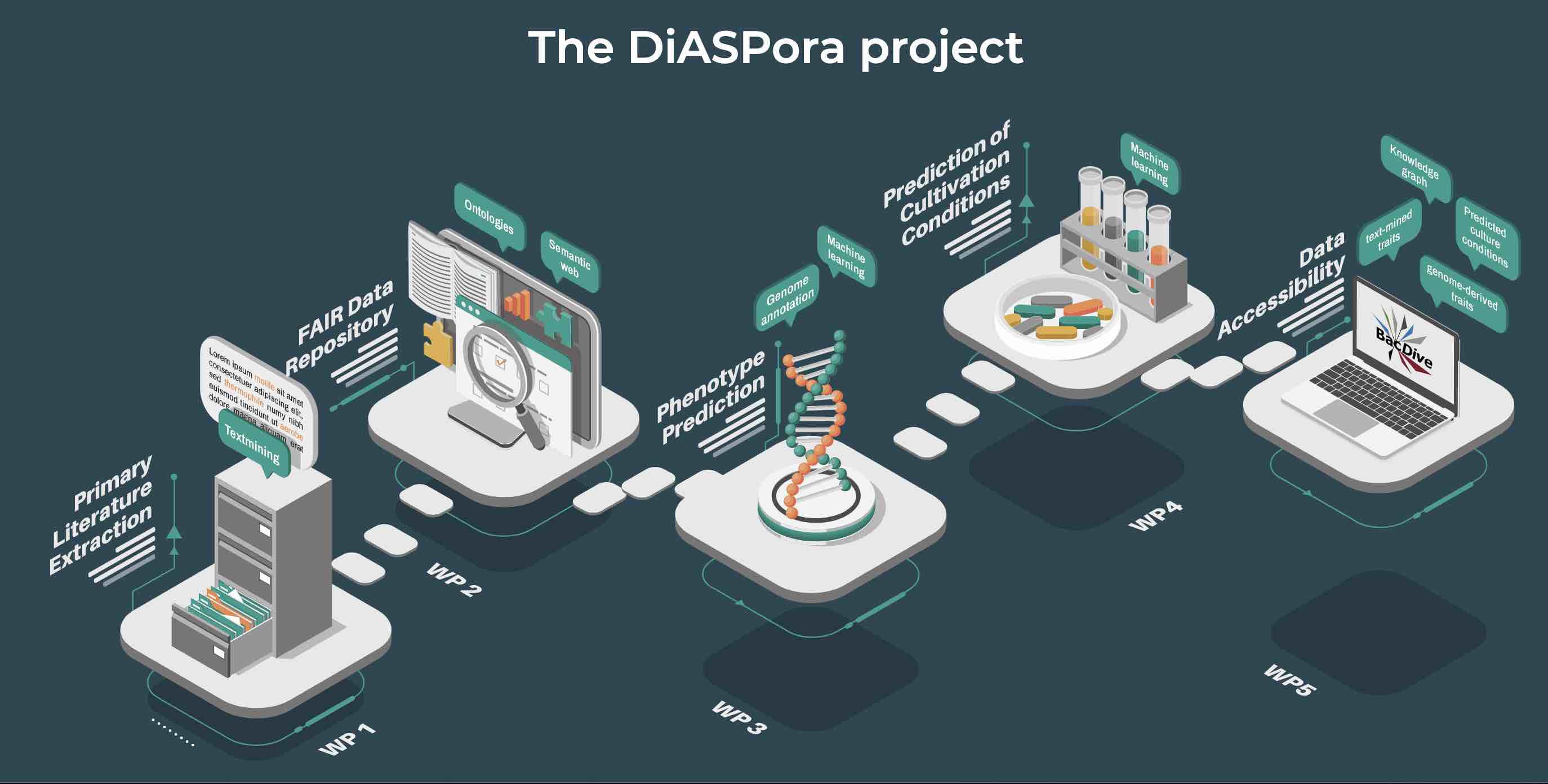
Source: https://diaspora-project.de/
Search projects by keywords:
-
The Hessian Center for Artificial Intelligence
Goesmann Group, Gießen University, Service Center: Bielefeld-Giessen Resource Center for Microbial Bioinformatics - BiGi
Established in 2020, the Hessian Center for Artificial Intelligence is a Germany-wide unique institution supported by 13 different universities bundling their expertise. In its five-year development phase, the state of Hesse is funding the center with a total amount of 38 million euros; 20 additional professorships are going to be established. The main center will be located at the TU Darmstadt, while a secondary site will be hosted by the Goethe University in Frankfurt. The center is complemented by additional regional locations at the participating institutions, among them the JLU Giessen. The AI center will offer study courses in all disciplines and also set up a graduate school in order to attract excellent scientists to Hesse. Artificial intelligence provides enormous scientific as well as economic potential, for example in agriculture or in the medical sector, facilitating the fast analysis of vast amounts of highly complex data.
At the JLU Giessen, Prof. Alexander Goesmann has already been involved in the founding phase of the new center, and an additional professorship dedicated to the subject area “Predictive Deep Learning in Medicine and Healthcare” is going to be established at the department of medicine, as well. Prof. Goesmann’s group routinely employs machine-learning techniques for the successful analysis of large biological data sets. They have established and continuously maintain an extensive cloud-based compute infrastructure comprising over 7,500 CPU cores and 16 PB storage capacity, also featuring dedicated GPU accelerators for high-throughput machine-learning purposes. The compute cloud is offered to German bioinformatics scientists free of charge, and is routinely employed for fast and efficient processing of biological data. In bioinformatics, many tools rely on established artificial intelligence approaches such as machine-learning, e.g. for the identification and classification of novel genes, and it is already foreseeable that the field will significantly benefit from the newly founded Hessian Center for Artificial Intelligence.
For further information, please visit the Hessian Center for Artificial Intelligence website. To access the de.NBI Cloud visitde.NBI Cloud.

Source: https://hessian.ai/
Search projects by Keywords:
