Overmann Group, Leibniz Institute DSMZ, Service center: Center for Biological Data – BioData
The rapidly increasing amount of data produced in science comes with new opportunities in analysis but also with new challenges in processing, interpreting, and storing data. The risk is high that large amounts of the data are only superficially analyzed and are being dumped on local storage without sufficient metadata enabling reuse of the data. Moreover, publication numbers rise quickly, but the data aggregated are difficult to access and not available for large scale analysis.
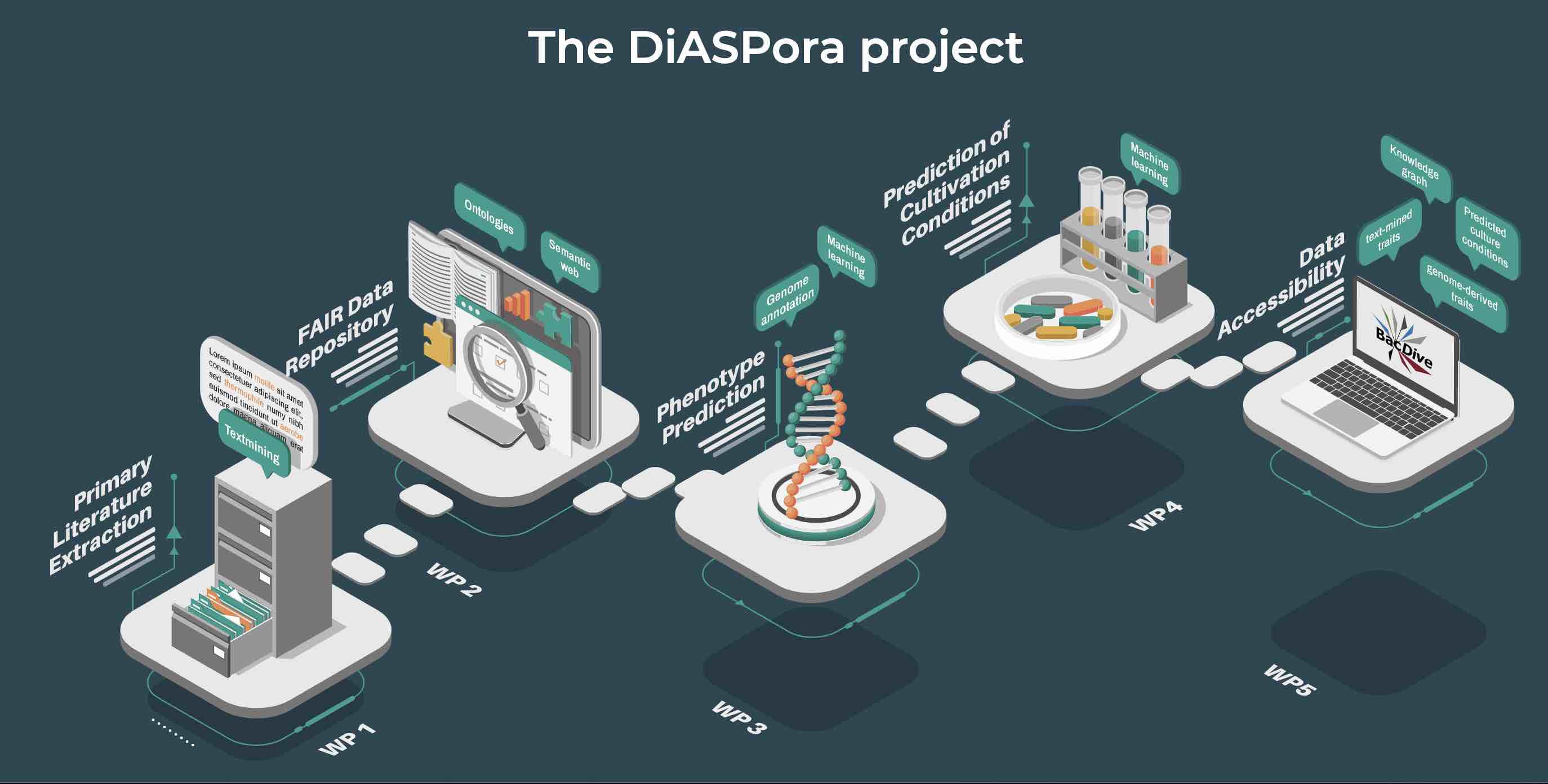
In a new approach (DiASPora project), the Leibniz Institute DSMZ is synthesizing information for bacterial species from diverse sources, together with the TIB Hanover, and ZB MED Cologne and publishes these in the database BacDive.
So far, the curation of scientific data is still a largely manual process that does not scale with the increasing amounts of data published every day. Therefore, workflows are developed that apply text mining combined with machine learning approaches including deep neural networks to automatically extract data for more than 150 microbial data types from literature. To attain high quality and correctness of the extracted data, a manual curation feedback loop will be integrated into the text mining pipeline, which enables to train the AI and thereby successively improve the quality.
The increasing availability of genomic data enables the prediction of bacterial traits based on genome annotation data. To achieve high quality in function prediction, classifiers are trained and tested with phenotypic data from BacDive and standardized genome annotations. An iterative optimization process including manual curation will retrieve the best models for a number of traits, allowing us to make predictions with a high confidence level for so far poorly studied bacteria.
All data that reach a high level of confidence will be standardized and published in BacDive (de.NBI tool) in a machine-interpretable format, ready for reuse and large-scale analysis.
For further information visit the following websites for DiASPora project and BacDive.
Source: https://diaspora-project.de/
Search projects by keywords:

