
Proteomics
-
DeePSIVal - Deep Learning approach to validation of spectrum identifications
Benndorf Group, Max Planck Institute for Dynamics of Complex Technical Systems, Magdeburg, Service Center: Bielefeld-Giessen Resource Center for Microbial Bioinformatics - BiGi
Proteomics is a field of molecular biology that targets improvements in medical, pharmaceutical and environmental research, and diagnostics. The computational analysis of the recorded mass spectrometry data has become one of the biggest challenges in the field of proteomics, in particular the problem of determining the validity of an identified peptide-spectrum match (PSM). Metaproteomics, which applies proteomics tools to microbial communities, faces further challenges regarding validation of PSMs.
Commonly employed methods for the validation of PSMs, like an FDR estimation with the Target-Decoy approach, rely heavily on expert systems and require the entirety of the data set at once to work. These methods are highly dependent on carefully designed scoring functions that use varying feature representations to achieve validation. Multiple issues arise, such as different outputs depending on method used and the inapplicability for streaming. In metaproteomics, protein database choice is an even bigger issue for PSM validation, because unlike in single-organism proteomics, protein databases are large to accommodate the uncertainty about the species present in a given sample, which in turn negatively influences PSM validation.
Our new implementation - Deep-learning Peptide Spectrum Identification Validator (DeePSIVal) - employs a deep learning architecture comprised of multiple GRU units organized in a many-to-one configuration to generate a multi-dimensional representation of the pairs of peptide sequences and spectra. A CNN union model interprets the tensor representation to correctly validate the encoded peptide spectrum match. Therefore, DeePSIVal relies on raw data and automatic feature detection. By using this method, PSM validation is independent of the protein database used, thus eliminating the effects of database size.
In our previous work, a machine learning approach was implemented and trained with prepared mass spectrometry data sets done for this specific purpose as well as data sets from the public database PRIDE. We will compare the DeePSIVal against other approaches with regard to model performance and time performance.
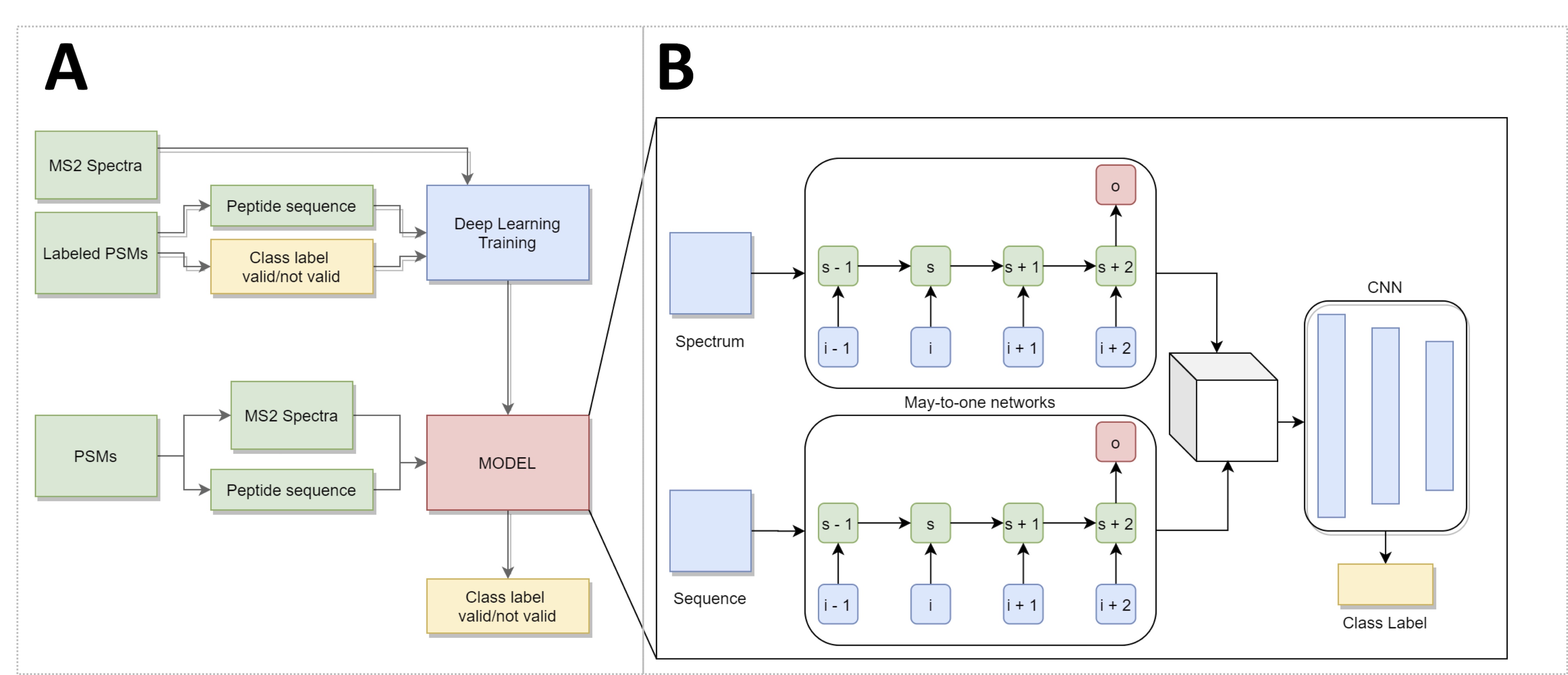
Workflow (A) and architecture (B) of the implemented DeePSIVal prototype. (A) MS2 spectra and the peptide sequence – the raw data for a peptide spectrum match – are used directly as input. For training a Class label has to be provided; the finished model can output a class label for new PSMs. (B) Deep learning architecture consists of 3 components. Two Many-to-One networks read the spectrum and sequence data of a PSM. The resulting output is concatenated and evaluated by a convolutional neural network, which will convert it into a class label.
Search projects by keywords:
-
DeProVIDEO - Deep Learning for Protein Variants Detection
Eisenacher Group, Ruhr University Bochum, Service center: Bioinformatics for Proteomics – BioInfra.Prot
The overarching goal of “DeProVIDEO – Deep Learning for Protein VarIants DEtectiOn” is the detection of variants in the amino acid sequence of proteins in mass spectrometry data. The complete concept can be split into two parts, which both base on deep learning approaches.
Genetic variants can be the cause of specific diseases or give a predisposition to these, while often proteins are the actually afflicted biomolecules of a genetic variation. Changes in the genome can cause simple amino acid exchanges in corresponding proteins, but could also lead to more complex changes like insertions or deletions of longer sequences. While the genetic causes for these protein variants are known, tools to detect variants without a known genetic background will be developed in DeProVIDEO.
As the insertion of variants into a given protein database drastically increases the search space, problems occur during a statistical analysis especially considering the estimation of false positive rates of identifications. Therefore, the first part of the project addresses the development of a spectrum centric, instead of a global, false positive estimation of identified peptides. This will be approached by the application of spectra predictions for database annotated peptide sequences and variants using a deep neural network.
The second part of the project uses specific deep learning algorithms to identify peptide sequences of measured spectra without database information by so called de novo strategies. With this approach yet unknown variants could be identified, which might originate neither from genetic variants nor could be detected by other proteomic methods.
All created tools and models will be made publicly available to the proteomics community. For further information visit the website of the Medical Proteome Center.
Search projects by keywords:
