
Drug Discovery
-
Identification of new antimicrobial resistance targets by high-throughput deep learning
Goesmann Group, Gießen University, Service Center: Bielefeld-Giessen Resource Center for Microbial Bioinformatics - BiGi
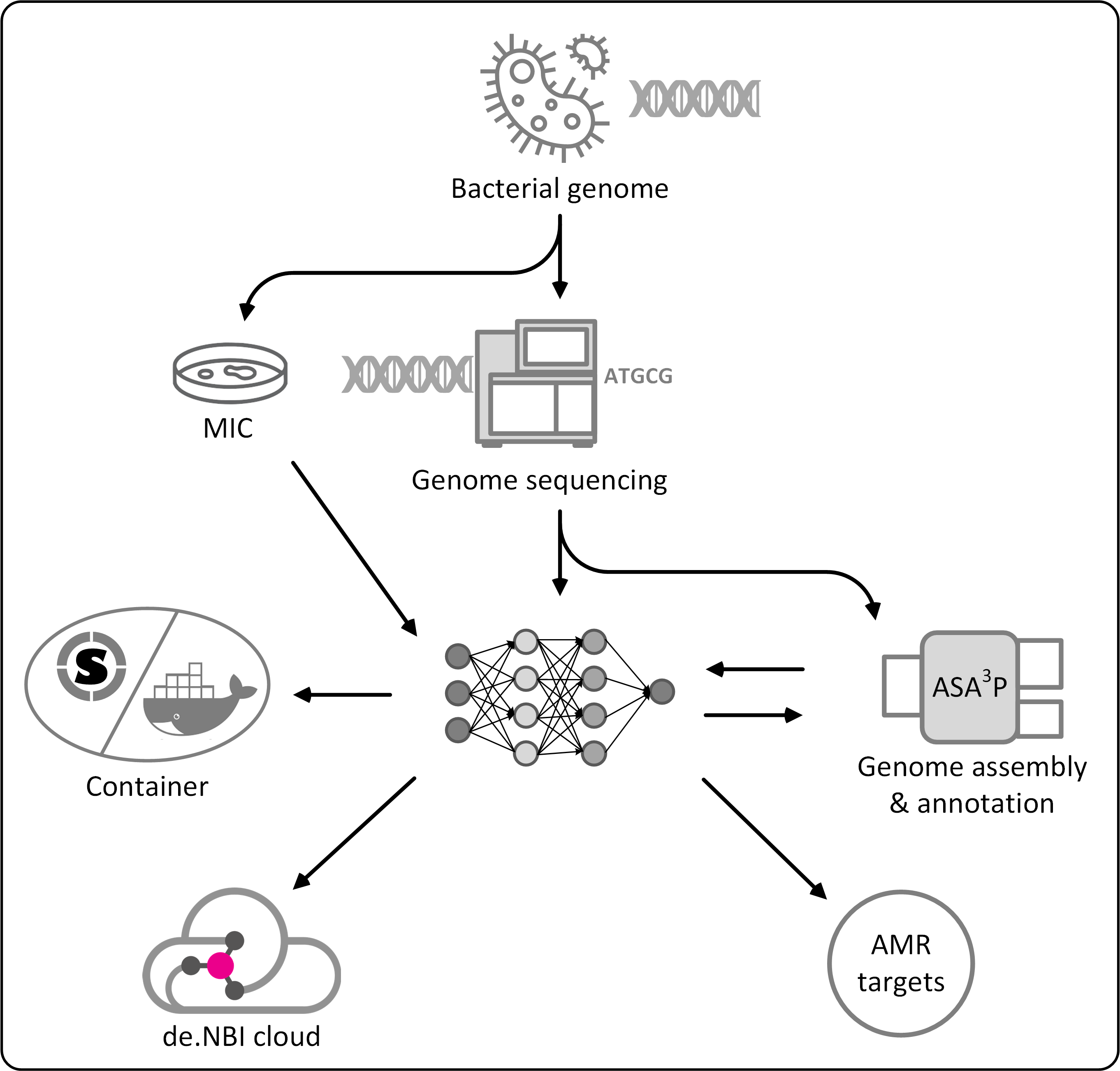
Antibiotic resistant bacteria have become a global severe threat for public health. Their surveillance and containment as well as the timely identification of new genetic determinants of antibiotic resistances are important tools to fight back emergent as well as established pathogens. Addressing these challenges, researchers from universities in Giessen and Marburg have started a new project called Deep-iAMR to combine expertise in medical microbiology, machine learning and cloud computing techniques. The aim of this new collaboration is to screen large amounts of genomic as well as phenotypic data for complex patterns between genomic and phenotypic data to find the genetic determinants of hard to detect or entirely new antimicrobial resistances. However, the application of modern artificial intelligence methodologies to find these patterns requires massive computing resources as well as specialized hardware, such as GPUs, tensors and FPGUs.
Researchers within the Deep-iAMR project take advantage of the cloud computing infrastructure of the de.NBI network which provides required computing resources as well as specialized hardware on demand. Resulting new methodologies and scalable tools will be shared with the scientific community and discovered new antibiotic targets will help to design necessary and urgently required new drugs to cure infections with highly-resistant pathogens.
For further information, please visit Deep-iAMR. To access the de.NBI Cloud visit de.NBI Cloud.
 Search projects by keywords:
Search projects by keywords: -
REmatch - Cloud AI for drug discovery
Boutros Group, DKFZ Heidelberg, Service center: Heidelberg Center for Human Bioinformatics – HD-HuB
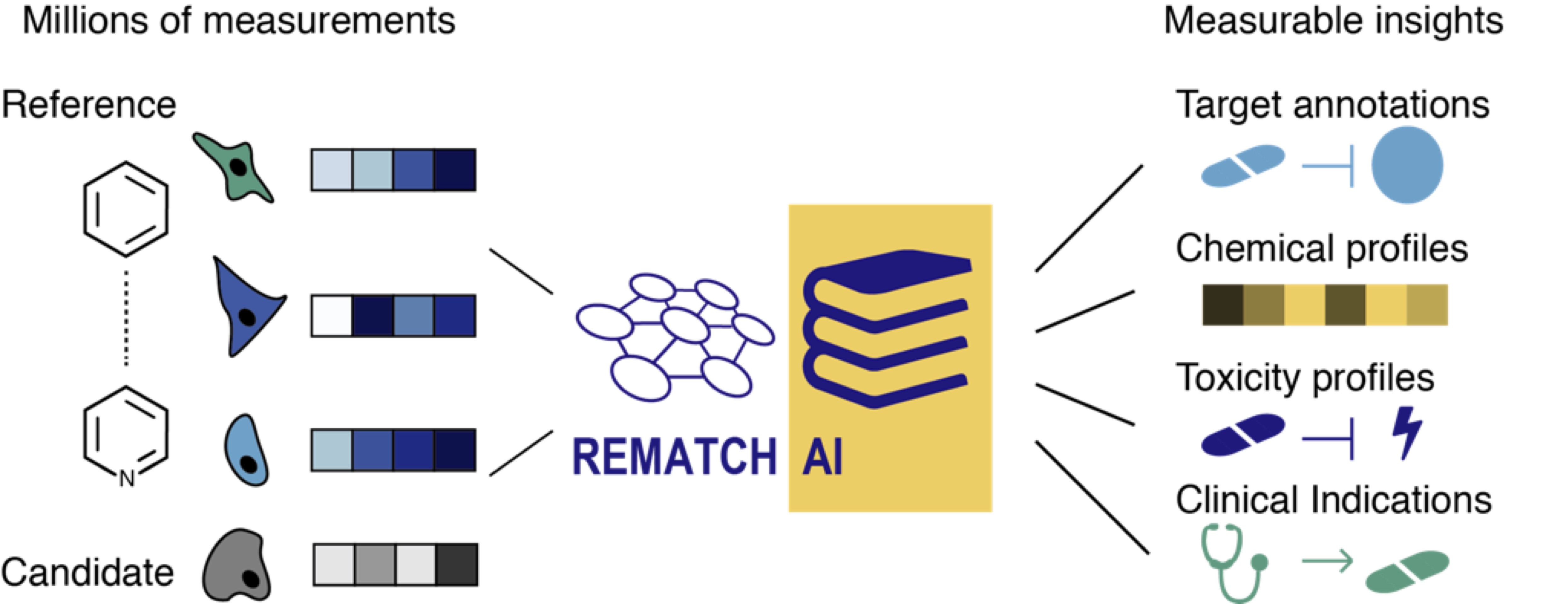
In the REmatch project we develop cloud technologies that combine advanced image analysis and an artificial intelligence (AI) to predict a drug’s mechanisms of action and its polypharmacology. By this means, REmatch aims to save resources, to avoid unnecessary complex tests, and to facilitate drug repurposing by identifying new applications for approved drugs.
High drug attrition rates, few discoveries of new drug targets and patent cliff together pose a major problem for drug development. Despite better technologies and a deeper understanding of biological mechanisms, the efficiency at which novel drugs reach the market steadily declines. Many candidates fail late in clinical development due to side-effects or the lack of a therapeutic window. In addition, many promising or approved drugs harbour unrecognized therapeutic efficacy in other indications that were not in the focus during initial development.
As part of the REmatch project, we develop machine learning strategies to compare drug responses of candidate drugs to a reference database containing the responses of thousands of approved drugs. This facilitates the profiling of side-effects such as toxicity for candidate drugs. By comparing the responses of known drugs, REmatch further allows to identify drugs which harbour potential for other indications than they were initially developed for. In our approach, the responses of candidates and known drugs are represented as profiles which are derived from images of cultured cells that were exposed to these drugs. We generate such images at low costs and in a highly parallel fashion using an automated microscopy platform. REmatch can be used early during the drug discovery process to flag candidates sharing e.g. known toxicity of marketed drugs. REmatch employs a cloud-based, scalable end-to-end image analysis to derive phenotypic profiles of drug treated cells in a highly efficient manner. This removes a major bottleneck arising when thousands to millions of images are collected. With REmatch we offer a solution that enables researchers to fast and cost-efficiently characterize candidate molecules during pre-clinical development.
REmatch is supported by an ERC Proof of Concept grant and as a part of the proof-of-concept evaluation we used the de.NBI Cloud to build a reference database that contains the responses of more than 2000 drugs in 20 cancer cell lines. The cloud resources were used to streamline the analysis of hundreds of thousands of images with the REmatch technology. REmatch delivers our analysis and data processing pipelines, as well as access to the reference database in any cloud. The concept of cloud-based analysis, know-how and infrastructure addresses an unmet need and makes a routinely employment of image-based profiling analysis in drug discovery commercially attractive for pharmaceutical companies and viable for academic labs.
For further information, visit the website of the Division of Signaling and Functional Genomics at DKFZ Heidelberg.
Search projects by keywords:
