Goesmann Group, Gießen University, Service Center: Bielefeld-Giessen Resource Center for Microbial Bioinformatics - BiGi
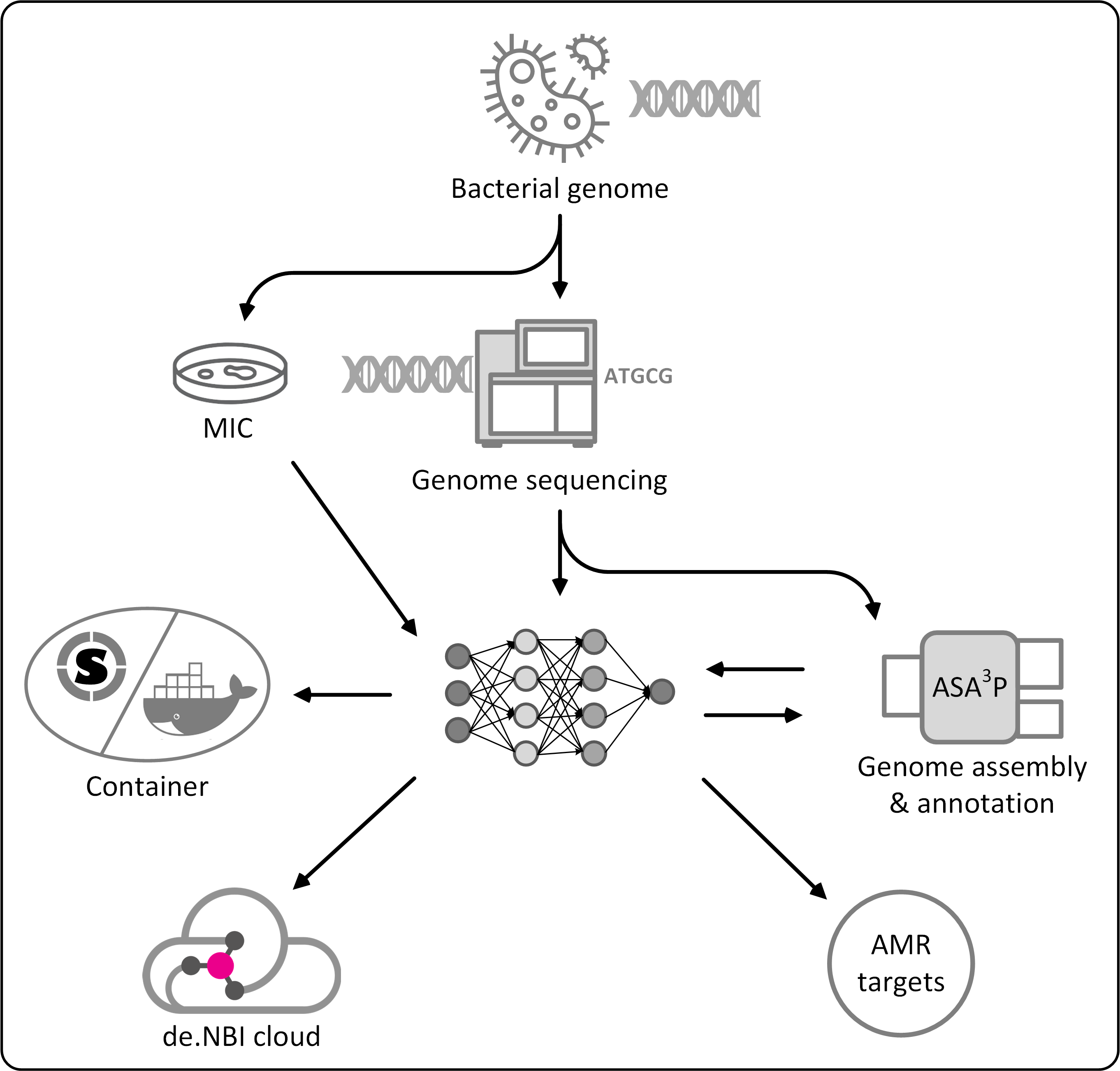
Antibiotic resistant bacteria have become a global severe threat for public health. Their surveillance and containment as well as the timely identification of new genetic determinants of antibiotic resistances are important tools to fight back emergent as well as established pathogens. Addressing these challenges, researchers from universities in Giessen and Marburg have started a new project called Deep-iAMR to combine expertise in medical microbiology, machine learning and cloud computing techniques. The aim of this new collaboration is to screen large amounts of genomic as well as phenotypic data for complex patterns between genomic and phenotypic data to find the genetic determinants of hard to detect or entirely new antimicrobial resistances. However, the application of modern artificial intelligence methodologies to find these patterns requires massive computing resources as well as specialized hardware, such as GPUs, tensors and FPGUs.
Researchers within the Deep-iAMR project take advantage of the cloud computing infrastructure of the de.NBI network which provides required computing resources as well as specialized hardware on demand. Resulting new methodologies and scalable tools will be shared with the scientific community and discovered new antibiotic targets will help to design necessary and urgently required new drugs to cure infections with highly-resistant pathogens.
For further information, please visit Deep-iAMR. To access the de.NBI Cloud visit de.NBI Cloud.
 Search projects by keywords:
Search projects by keywords:

