
Phenotype
-
LAMarCK - LongitudinAI Multiomics Characterization of disease using prior Knowledge
Zeller Group, Saez-Rodriguez Group, Schlesner Group, Bork Group, and Korbel Group, Service center: Heidelberg Center for Human Bioinformatics – HD-HuB
The need for proper integration of molecular omics data from human individuals is arguably one of the biggest challenges in current biomedical research, as these data cover different functional aspects that facilitate basic research, but also reveal differences between human physiological states with medical relevance (e.g. disease versus healthy, different disease subtypes etc.). To tackle this challenge, we formed a team of researchers with complementary areas of expertise from different Heidelberg institutions, all associated with the de.NBI node HD-HuB, which delivers services on human genomics and human microbiomics. Both these fields rely on similar kinds of data, which are increasingly coming from longitudinal studies assessing multiple omics approaches. Thus, they face similar technical and methodological challenges - and closer communication, as well as integrative approaches bridging both fields hold great promise for improved diagnostic, prognostic and therapeutic approaches, as is already becoming apparent for type-2-diabetes onset.
The LAMarCK consortium on the one hand aims to devise generic approaches to integrate heterogeneous longitudinal multi-omics data across microbiome and host, on the other hand it will also develop frameworks that make use of domain-specific prior knowledge on the underlying biological mechanisms for improved interpretability of the results. Funded by the BMBF (grant no. 031L0181A), the consortium is applying and benchmarking state-of-the-art machine learning approaches for longitudinal multi-omics data from microbial and host cells and augmenting these by integration with biomolecular knowledge bases. These new approaches will help dissect the molecular mechanisms underlying human diseases, such as colorectal cancer.
Search projects by Keywords:
-
Machine Learning in translational (single-cell) omics
Group for Clinical bioinformatics & Machine learning in translational single-cell biology, University Hospital Tübingen, University Tübingen, Service Center: Center for Integrative Bioinformatics - CiBi
Cell subpopulations play a pivotal role in the initiation and progression of immune processes and complex diseases. We plan to perform CPU/GPU based computations to elucidate intra- and intercellular mechanisms of such cell subsets conferring their function and association to organism-level phenotypes. Specifically, computations will be performed for development to leverage single-cell measurement based studies to this end. Specifically, we developed machine learning approaches to identify rare, phenotype associated cell populations either via deep or shallow unsupervised or supervised learning. High-dimensional single-cell snapshot measurements are now increasingly utilized to study dynamic processes, complementing our recent efforts based on time-lapse measurements. However, it is not trivial to derive interpretable dynamic models from these. To this end we developed and will further benchmark sparse regression approaches, probabilistic graphical modeling, supervised surrogate learning approaches for large scale reaction network inference and the Dynamic Distribution Decomposition, as well as psupertime, a supervised pseudotime approach for single-cell RNA seq time series based on a regression model, which explicitly uses time series labels as input. Methods will be benchmarked on both unsensitive research data, as well as applied on possibly sensitive patient data from collaborations with partners at the University/University Hospital Tübingen. Initial activity in this project involved simulation and AI based inference of T cell exhaustion differentiation in chronic infections (see Figure).
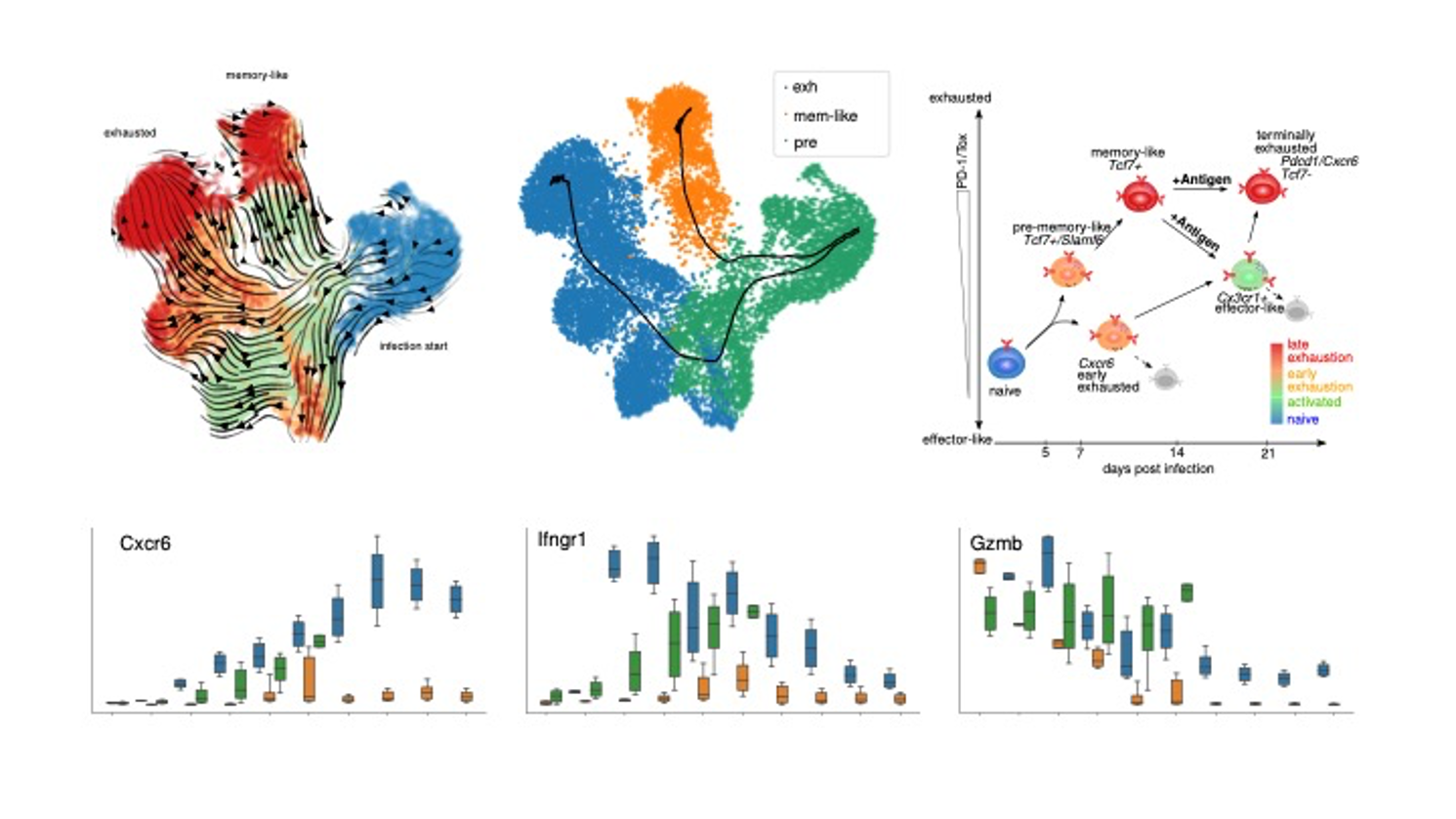
Simulation and AI based inference of T cell exhaustion differentiation in chronic infections. Dynamic and Markov Chain modeling of cellular bifurcated differentiation processes including classification based molecular characterization of individual differentiation branches. These analyses and follow up experiments (data not shown) resulted in high level differentiation model (Cerletti et al., in prep.).
Search projects by keywords:
