Group for Clinical bioinformatics & Machine learning in translational single-cell biology, University Hospital Tübingen, University Tübingen, Service Center: Center for Integrative Bioinformatics - CiBi
Cell subpopulations play a pivotal role in the initiation and progression of immune processes and complex diseases. We plan to perform CPU/GPU based computations to elucidate intra- and intercellular mechanisms of such cell subsets conferring their function and association to organism-level phenotypes. Specifically, computations will be performed for development to leverage single-cell measurement based studies to this end. Specifically, we developed machine learning approaches to identify rare, phenotype associated cell populations either via deep or shallow unsupervised or supervised learning. High-dimensional single-cell snapshot measurements are now increasingly utilized to study dynamic processes, complementing our recent efforts based on time-lapse measurements. However, it is not trivial to derive interpretable dynamic models from these. To this end we developed and will further benchmark sparse regression approaches, probabilistic graphical modeling, supervised surrogate learning approaches for large scale reaction network inference and the Dynamic Distribution Decomposition, as well as psupertime, a supervised pseudotime approach for single-cell RNA seq time series based on a regression model, which explicitly uses time series labels as input. Methods will be benchmarked on both unsensitive research data, as well as applied on possibly sensitive patient data from collaborations with partners at the University/University Hospital Tübingen. Initial activity in this project involved simulation and AI based inference of T cell exhaustion differentiation in chronic infections (see Figure).
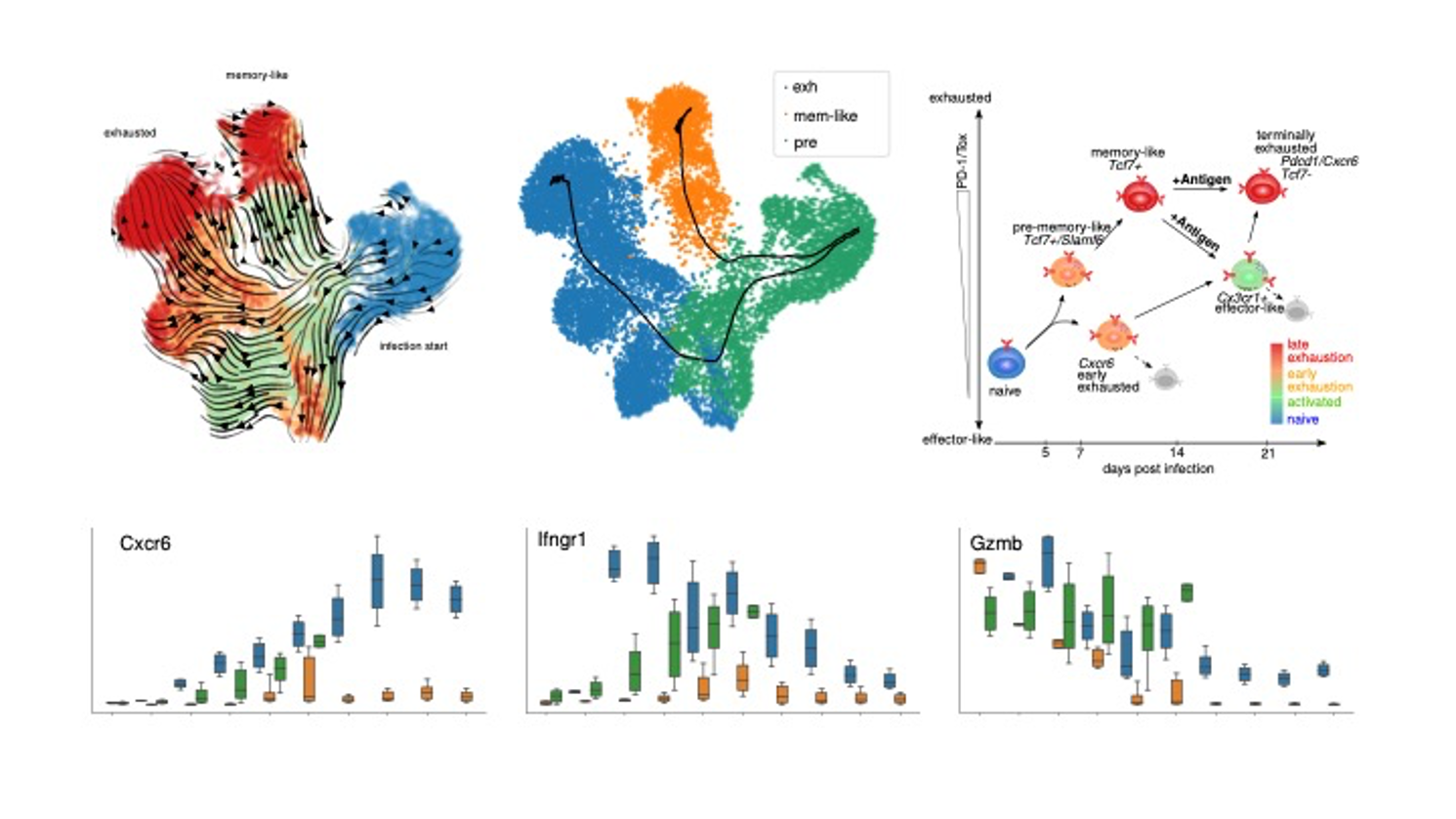
Simulation and AI based inference of T cell exhaustion differentiation in chronic infections. Dynamic and Markov Chain modeling of cellular bifurcated differentiation processes including classification based molecular characterization of individual differentiation branches. These analyses and follow up experiments (data not shown) resulted in high level differentiation model (Cerletti et al., in prep.).
Search projects by keywords:

